The 1.6-A crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad
- PMID: 12621151
- PMCID: PMC152259
- DOI: 10.1073/pnas.0530312100
The 1.6-A crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad
Abstract
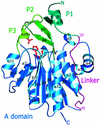
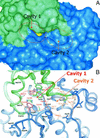
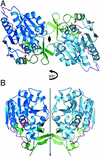
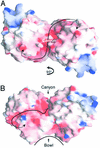
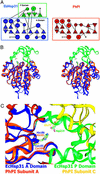
Heat shock proteins (Hsps) play essential protective roles under stress conditions by preventing the formation of protein aggregates and degrading misfolded proteins. EcHsp31, the yedU (hchA) gene product, is a representative member of a family of chaperones that alleviates protein misfolding by interacting with early unfolding intermediates. The 1.6-A crystal structure of the EcHsp31 dimer reveals a system of hydrophobic patches, canyons, and grooves, which may stabilize partially unfolded substrate. The presence of a well conserved, yet buried, triad in each two-domain subunit suggests a still unproven hydrolytic function of the protein. A flexible extended linker between the A and P domains may play a role in conformational flexibility and substrate binding. The alpha-beta sandwich of the EcHsp31 monomer shows structural similarity to PhPI, a protease belonging to the DJ-1 superfamily. The structure-guided sequence alignment indicates that Hsp31 homologs can be divided in three classes based on variations in the P domain that dramatically affect both oligomerization and catalytic triad formation.
Figures

Similar articles
-
The crystal structure of Escherichia coli heat shock protein YedU reveals three potential catalytic active sites.Protein Sci. 2003 Oct;12(10):2303-11. doi: 10.1110/ps.03121403. Protein Sci. 2003. PMID: 14500888 Free PMC article.
-
A new native EcHsp31 structure suggests a key role of structural flexibility for chaperone function.Protein Sci. 2004 Jan;13(1):269-77. doi: 10.1110/ps.03399604. Protein Sci. 2004. PMID: 14691241 Free PMC article.
-
Structural and biochemical studies on Vibrio cholerae Hsp31 reveals a novel dimeric form and Glutathione-independent Glyoxalase activity.PLoS One. 2017 Feb 24;12(2):e0172629. doi: 10.1371/journal.pone.0172629. eCollection 2017. PLoS One. 2017. PMID: 28235098 Free PMC article.
-
The structural basis of mode of activation and functional diversity: a case study with HtrA family of serine proteases.Arch Biochem Biophys. 2011 Dec 15;516(2):85-96. doi: 10.1016/j.abb.2011.10.007. Epub 2011 Oct 18. Arch Biochem Biophys. 2011. PMID: 22027029 Review.
-
[Bacterial ClpX protease structure and function--a review].Wei Sheng Wu Xue Bao. 2010 Oct;50(10):1281-7. Wei Sheng Wu Xue Bao. 2010. PMID: 21141460 Review. Chinese.
Cited by
-
Dissection of the dimerization modes in the DJ-1 superfamily.Mol Cells. 2012 Feb;33(2):163-71. doi: 10.1007/s10059-012-2220-6. Epub 2012 Jan 2. Mol Cells. 2012. PMID: 22228183 Free PMC article.
-
The crystal structure of Escherichia coli heat shock protein YedU reveals three potential catalytic active sites.Protein Sci. 2003 Oct;12(10):2303-11. doi: 10.1110/ps.03121403. Protein Sci. 2003. PMID: 14500888 Free PMC article.
-
Cyclosporine A Treatment Abrogates Ischemia-Induced Neuronal Cell Death by Preserving Mitochondrial Integrity through Upregulation of the Parkinson's Disease-Associated Protein DJ-1.CNS Neurosci Ther. 2016 Jul;22(7):602-10. doi: 10.1111/cns.12546. Epub 2016 Jun 1. CNS Neurosci Ther. 2016. PMID: 27247192 Free PMC article.
-
De Novo Transcriptome of Mammillaria bombycina (Cactaceae) under In Vitro Conditions and Identification of Glyoxalase Genes.Plants (Basel). 2022 Jan 31;11(3):399. doi: 10.3390/plants11030399. Plants (Basel). 2022. PMID: 35161380 Free PMC article.
-
Experimental and computational studies indicate the mutation of Glu12 to increase the thermostability of oligomeric protease from Pyrococcus horikoshii.J Mol Model. 2011 Jun;17(6):1241-9. doi: 10.1007/s00894-010-0819-0. Epub 2010 Aug 15. J Mol Model. 2011. PMID: 20711794
References
-
- Hartl F U, Hayer-Hartl M. Science. 2002;295:1852–1858. - PubMed
-
- Ben-Zvi A P, Goloubinoff P. J Struct Biol. 2001;135:84–93. - PubMed
-
- Gottesman S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. - PubMed
-
- Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. - PubMed
-
- Spiess C, Beil A, Ehrmann M. Cell. 1999;97:339–347. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

