RPA is an initiation factor for human chromosomal DNA replication
- PMID: 12626714
- PMCID: PMC152871
- DOI: 10.1093/nar/gkg269
RPA is an initiation factor for human chromosomal DNA replication
Abstract
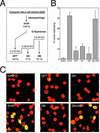
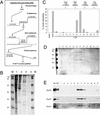
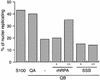
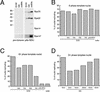
The initiation of chromosomal DNA replication in human cell nuclei is not well understood because of its complexity. To allow investigation of this process on a molecular level, we have recently established a cell-free system that initiates chromosomal DNA replication in an origin-specific manner under cell cycle control in isolated human cell nuclei. We have now used fractionation and reconstitution experiments to functionally identify cellular factors present in a human cell extract that trigger initiation of chromosomal DNA replication in this system. Initial fractionation of a cytosolic extract indicates the presence of at least two independent and non-redundant initiation factors. We have purified one of these factors to homogeneity and identified it as the single-stranded DNA binding protein RPA. The prokaryotic single-stranded DNA binding protein SSB cannot substitute for RPA in the initiation of human chromosomal DNA replication. Antibodies specific for human RPA inhibit the initiation step of human chromosomal DNA replication in vitro. RPA is recruited to DNA replication foci and becomes phosphorylated concomitant with the initiation step in vitro. These data establish a direct functional role for RPA as an essential factor for the initiation of human chromosomal DNA replication.
Figures

Similar articles
-
Distinct populations of human PCNA are required for initiation of chromosomal DNA replication and concurrent DNA repair.Exp Cell Res. 2005 Dec 10;311(2):240-50. doi: 10.1016/j.yexcr.2005.09.009. Epub 2005 Oct 14. Exp Cell Res. 2005. PMID: 16226749
-
Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex.Exp Cell Res. 2000 Feb 1;254(2):321-7. doi: 10.1006/excr.1999.4770. Exp Cell Res. 2000. PMID: 10640430
-
UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein.Mol Biol Cell. 2001 May;12(5):1199-213. doi: 10.1091/mbc.12.5.1199. Mol Biol Cell. 2001. PMID: 11359916 Free PMC article.
-
Replication protein A phosphorylation and the cellular response to DNA damage.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1015-24. doi: 10.1016/j.dnarep.2004.03.028. DNA Repair (Amst). 2004. PMID: 15279788 Review.
-
Eukaryotic single-stranded DNA binding proteins: central factors in genome stability.Subcell Biochem. 2010;50:143-63. doi: 10.1007/978-90-481-3471-7_8. Subcell Biochem. 2010. PMID: 20012581 Review.
Cited by
-
Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis.Mol Cell Biol. 2004 Apr;24(7):2853-62. doi: 10.1128/MCB.24.7.2853-2862.2004. Mol Cell Biol. 2004. PMID: 15024074 Free PMC article.
-
Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication.PLoS One. 2010 Oct 27;5(10):e13673. doi: 10.1371/journal.pone.0013673. PLoS One. 2010. PMID: 21060685 Free PMC article.
-
A conserved motif of vertebrate Y RNAs essential for chromosomal DNA replication.RNA. 2009 Jul;15(7):1375-85. doi: 10.1261/rna.1472009. Epub 2009 May 27. RNA. 2009. PMID: 19474146 Free PMC article.
-
Visualization of bidirectional initiation of chromosomal DNA replication in a human cell free system.Nucleic Acids Res. 2005 Dec 6;33(21):6931-41. doi: 10.1093/nar/gki994. Print 2005. Nucleic Acids Res. 2005. PMID: 16332696 Free PMC article.
-
Functional requirement of noncoding Y RNAs for human chromosomal DNA replication.Mol Cell Biol. 2006 Sep;26(18):6993-7004. doi: 10.1128/MCB.01060-06. Mol Cell Biol. 2006. PMID: 16943439 Free PMC article.
References
-
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. - PubMed
-
- Diffley J.F. and Labib,K. (2002) The chromosome replication cycle. J. Cell Sci., 115, 869–872. - PubMed
-
- Lei M. and Tye,B.K. (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci., 114, 1447–1454. - PubMed
-
- Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

