Transcriptional network controlled by the trithorax-group gene ash2 in Drosophila melanogaster
- PMID: 12626737
- PMCID: PMC152285
- DOI: 10.1073/pnas.0538075100
Transcriptional network controlled by the trithorax-group gene ash2 in Drosophila melanogaster
Erratum in
- Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):17141
Abstract
The transcription factor absent, small, or homeotic discs 2 (ash2) gene is a member of the trithorax group of positive regulators of homeotic genes. Mutant alleles for ash2 are larvalpupal lethals and display imaginal disc and brain abnormalities. The allele used in this study is a true mutant for the trithorax function and lacks the longest transcript present in wild-type flies. In an attempt to identify gene targets of ash2, we have performed an expression analysis by using cDNA microarrays. Genes involved in cell cycle, cell proliferation, and cell adhesion are among these targets, and some of them are validated by functional and expression studies. Even though trithorax proteins act by modulating chromatin structure at particular chromosomal locations, evidence of physical aggregation of ash2-regulated genes has not been found. This work represents the first microarray analysis, to our knowledge, of a trithorax-group gene.
Figures
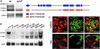
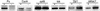
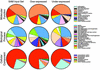


References
-
- Simon J. Curr Opin Cell Biol. 1995;7:376–385. - PubMed
-
- Schumacher A, Magnuson T. Trends Genet. 1997;13:167–170. - PubMed
-
- Kennison J A. Annu Rev Genet. 1995;29:289–303. - PubMed
-
- Mahmoudi T, Verrijzer C P. Oncogene. 2001;20:3055–3066. - PubMed
-
- Francis N J, Kingston R E. Nat Rev Mol Cell Biol. 2001;2:409–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

