Integrating regulatory motif discovery and genome-wide expression analysis
- PMID: 12626739
- PMCID: PMC152294
- DOI: 10.1073/pnas.0630591100
Integrating regulatory motif discovery and genome-wide expression analysis
Abstract
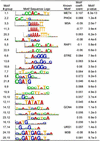
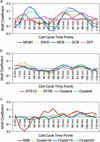
We propose motif regressor for discovering sequence motifs upstream of genes that undergo expression changes in a given condition. The method combines the advantages of matrix-based motif finding and oligomer motif-expression regression analysis, resulting in high sensitivity and specificity. motif regressor is particularly effective in discovering expression-mediating motifs of medium to long width with multiple degenerate positions. When applied to Saccharomyces cerevisiae, motif regressor identified the ROX1 and YAP1 motifs from Rox1p and Yap1p overexpression experiments, respectively; predicted that Gcn4p may have increased activity in YAP1 deletion mutants; reported a group of motifs (including GCN4, PHO4, MET4, STRE, USR1, RAP1, M3A, and M3B) that may mediate the transcriptional response to amino acid starvation; and found all of the known cell-cycle regulation motifs from 18 expression microarrays over two cell cycles.
Figures


References
-
- van Helden J, Andre B, Collado-Vides J. J Mol Biol. 1998;281:827–842. - PubMed
-
- Vilo J, Brazma A, Jonassen I, Robinson A, Ukkonen E. Proc Int Conf Intell Syst Mol Biol. 2000;8:384–394. - PubMed
-
- Sinha S, Tompa M. Proc Int Conf Intell Syst Mol Biol. 2000;8:344–354. - PubMed
-
- Hampson S, Kibler D, Baldi P. Bioinformatics. 2002;18:513–528. - PubMed
-
- Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Science. 1993;262:208–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

