Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae
- PMID: 12626741
- PMCID: PMC152254
- DOI: 10.1073/pnas.0634629100
Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae
Abstract
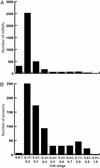
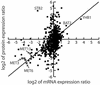
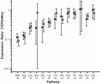
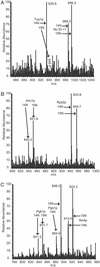
The mRNA and protein expression in Saccharomyces cerevisiae cultured in rich or minimal media was analyzed by oligonucleotide arrays and quantitative multidimensional protein identification technology. The overall correlation between mRNA and protein expression was weakly positive with a Spearman rank correlation coefficient of 0.45 for 678 loci. To place the data sets in a proper biological context, a clustering approach based on protein pathways and protein complexes was implemented. Protein expression levels were transcriptionally controlled for not only single loci but for entire protein pathways (e.g., Met, Arg, and Leu biosynthetic pathways). In contrast, the protein expression of loci in several protein complexes (e.g., SPT, COPI, and ribosome) was posttranscriptionally controlled. The coupling of the methods described provided insight into the biology of S. cerevisiae and a clustering strategy by which future studies should be based.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

