Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin
- PMID: 12627940
- PMCID: PMC1435692
- DOI: 10.1021/bi027224+
Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin
Abstract
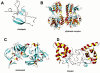
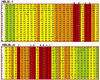
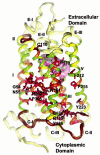
G-protein-coupled receptors (GPCRs) constitute a large superfamily of receptor proteins responsible for signal transduction (see
Figures

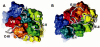


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

