Protein:protein interactions and the pairing of boundary elements in vivo
- PMID: 12629048
- PMCID: PMC196003
- DOI: 10.1101/gad.1052003
Protein:protein interactions and the pairing of boundary elements in vivo
Abstract
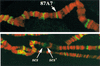
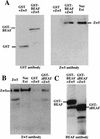
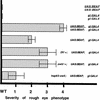
Although it is now well-established that boundary elements/insulators function to subdivide eukaryotic chromosomes into autonomous regulatory domains, the underlying mechanisms remain elusive. One idea is that boundaries act as barriers, preventing the processive spreading of "active" or "silenced" chromatin between domains. Another is that the partitioning into autonomous functional units is a consequence of an underlying structural subdivision of the chromosome into higher order "looped" domains. In this view, boundaries are thought to delimit structural domains by interacting with each other or with some other nuclear structure. The studies reported here provide support for the looped domain model. We show that the Drosophila scs and scs' boundary proteins, Zw5 and BEAF, respectively, interact with each other in vitro and in vivo. Moreover, consistent with idea that this protein:protein interaction might facilitate pairing of boundary elements, we find that that scs and scs' are in close proximity to each other in Drosophila nuclei.
Figures



References
-
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. - PubMed
-
- Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. - PubMed
-
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
