Biallelic germline transcription at the kappa immunoglobulin locus
- PMID: 12629064
- PMCID: PMC2193856
- DOI: 10.1084/jem.20021392
Biallelic germline transcription at the kappa immunoglobulin locus
Abstract
Rearrangement of antigen receptor genes generates a vast array of antigen receptors on lymphocytes. The establishment of allelic exclusion in immunoglobulin genes requires differential treatment of the two sequence identical alleles. In the case of the kappa immunoglobulin locus, changes in chromatin structure, methylation, and replication timing of the two alleles are all potentially involved in regulating rearrangement. Additionally, germline transcription of the kappa locus which precedes rearrangement has been proposed to reflect an opening of the chromatin structure rendering it available for rearrangement. As the initial restriction of rearrangement to one allele is critical to the establishment of allelic exclusion, a key question is whether or not germline transcription at the kappa locus is monoallelic or biallelic. We have used a sensitive reverse transcription-polymerase chain reaction (RT-PCR) assay and an RNA-fluorescence in situ hybridization (FISH) to show that germline transcription of the kappa locus is biallelic in wild-type immature B cells and in recombination activating gene (RAG)-/-, mu+ B cells. Therefore, germline transcription is unlikely to dictate which allele will be rearranged first and rather reflects a general opening on both alleles that must be accompanied by a mechanism allowing one of the two alleles to be rearranged first.
Figures



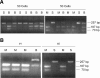
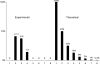

References
-
- Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581. - PubMed
-
- Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. - PubMed
-
- Schatz, D.G., M.A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell. 59:1035–1048. - PubMed
-
- Oettinger, M.A., D.G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 248:1517–1523. - PubMed
-
- Gellert, M. 1996. A new view of V(D)J recombination. Genes Cells. 1:269–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

