Leukemia inhibitory factor is a key signal for injury-induced neurogenesis in the adult mouse olfactory epithelium
- PMID: 12629183
- PMCID: PMC6741956
- DOI: 10.1523/JNEUROSCI.23-05-01792.2003
Leukemia inhibitory factor is a key signal for injury-induced neurogenesis in the adult mouse olfactory epithelium
Abstract
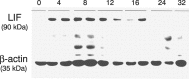
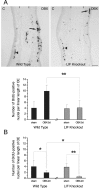
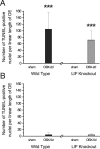
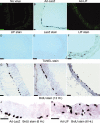
The mammalian olfactory epithelium (OE) is composed of primary olfactory sensory neurons (OSNs) that are renewed throughout adulthood by local, restricted neuronal progenitor cells. The molecular signals that control this neurogenesis in vivo are unknown. Using olfactory bulb ablation (OBX) in adult mice to trigger synchronous mitotic stimulation of neuronal progenitors in the OE, we show the in vivo involvement of a cytokine in the cellular events leading to the regeneration of the OE. We find that, of many potential mitogenic signals, only leukemia inhibitory factor (LIF) is induced before the onset of neuronal progenitor proliferation. The rise in LIF mRNA expression peaks at 8 hr after OBX, and in situ RT-PCR and immunocytochemistry indicate that LIF is upregulated, in part, in the injured neurons themselves. This rise in LIF is necessary for injury-induced neurogenesis, as OBX in the LIF knock-out mouse fails to stimulate cell proliferation in the OE. Moreover, delivery of exogenous LIF to the intact adult OE using an adenoviral vector stimulates BrdU labeling in the apical OE. Taken together, these results suggest that injured OSNs release LIF as a stimulus to initiate their own replacement.
Figures



References
-
- Banner LR, Moayeri NN, Patterson PH. Leukemia inhibitory factor is expressed in astrocytes following cortical brain injury. Exp Neurol. 1997;147:1–9. - PubMed
-
- Bauer S, Mauduit C, Jourdan F, Benahmed M, Moyse E. In vivo involvement of the cytokine LIF during lesion-induced renewal of olfactory sensory neurons in adult mouse. In: Patterson PH, Kordon C, Christen Y, editors. Neuro-immune interactions in neurologic and psychiatric disorders. Springer; Fondation IPSEN. Berlin: 2000. pp. 153–160.
-
- Buckland ME, Cunningham AM. Alterations in expression of the neurotrophic factors glial cell line-derived neurotrophic factor, ciliary neurotrophic factor and brain-derived neurotrophic factor, in the target-deprived olfactory neuroepithelium. Neuroscience. 1999;90:333–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
