The matrix attachment region in the Chinese hamster dihydrofolate reductase origin of replication may be required for local chromatid separation
- PMID: 12629222
- PMCID: PMC152283
- DOI: 10.1073/pnas.0437791100
The matrix attachment region in the Chinese hamster dihydrofolate reductase origin of replication may be required for local chromatid separation
Abstract
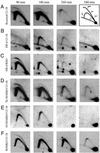
Centered in the Chinese hamster dihydrofolate reductase origin of replication is a prominent nuclear matrix attachment region (MAR). Indirect lines of evidence suggested that this MAR might be required for origin activation in early S phase. To test this possibility, we have deleted the MAR from a Chinese hamster ovary variant harboring a single copy of the dihydrofolate reductase locus. However, 2D gel replicon mapping shows that removal of the MAR has no significant effect either on the frequency or timing of initiation in this locus. Rather, fluorescence in situ hybridization studies on cells swollen under either neutral or alkaline conditions show that deletion of the MAR interferes with local separation of daughter chromatids. This surprising result provides direct genetic evidence that at least a subset of MARs performs an important biological function, possibly related to chromatid cohesion and separation.
Figures
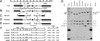


Similar articles
-
The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity.Mol Cell Biol. 2001 Feb;21(4):1098-110. doi: 10.1128/MCB.21.4.1098-1110.2001. Mol Cell Biol. 2001. PMID: 11158297 Free PMC article.
-
Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells.Mol Cell Biol. 1988 Dec;8(12):5398-409. doi: 10.1128/mcb.8.12.5398-5409.1988. Mol Cell Biol. 1988. PMID: 3244360 Free PMC article.
-
The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units.Mol Cell Biol. 2006 Feb;26(3):1051-62. doi: 10.1128/MCB.26.3.1051-1062.2006. Mol Cell Biol. 2006. PMID: 16428457 Free PMC article.
-
Mammalian origins of replication.Bioessays. 1992 Oct;14(10):651-9. doi: 10.1002/bies.950141002. Bioessays. 1992. PMID: 1365877 Review.
-
Sequence and context effects on origin function in mammalian cells.J Cell Biochem. 1996 Aug;62(2):210-22. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C210::AID-JCB9%3E3.0.CO;2-V. J Cell Biochem. 1996. PMID: 8844401 Review.
Cited by
-
The human beta-globin replication initiation region consists of two modular independent replicators.Mol Cell Biol. 2004 Apr;24(8):3373-86. doi: 10.1128/MCB.24.8.3373-3386.2004. Mol Cell Biol. 2004. PMID: 15060158 Free PMC article.
-
A revisionist replicon model for higher eukaryotic genomes.J Cell Biochem. 2008 Oct 1;105(2):321-9. doi: 10.1002/jcb.21828. J Cell Biochem. 2008. PMID: 18680119 Free PMC article. Review.
-
Repeat induces not only gene silencing, but also gene activation in mammalian cells.PLoS One. 2020 Jun 24;15(6):e0235127. doi: 10.1371/journal.pone.0235127. eCollection 2020. PLoS One. 2020. PMID: 32579599 Free PMC article.
-
Anchoring the genome.Genome Biol. 2008 Jan 22;9(1):201. doi: 10.1186/gb-2008-9-1-201. Genome Biol. 2008. PMID: 18226181 Free PMC article. Review.
-
In vivo mapping of arabidopsis scaffold/matrix attachment regions reveals link to nucleosome-disfavoring poly(dA:dT) tracts.Plant Cell. 2014 Jan;26(1):102-20. doi: 10.1105/tpc.113.121194. Epub 2014 Jan 31. Plant Cell. 2014. PMID: 24488963 Free PMC article.
References
-
- Marsden M P, Laemmli U K. Cell. 1979;17:849–858. - PubMed
-
- Saitoh Y, Laemmli U K. Cell. 1994;76:609–622. - PubMed
-
- Boveri T. Arch Zellforschung. 1909;3:181–191.
-
- Cremer T, Kreth G, Koester H, Fink R H, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Crit Rev Eukaryotic Gene Expression. 2000;10:179–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

