Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants
- PMID: 12631715
- PMCID: PMC151571
- DOI: 10.1091/mbc.e02-10-0633
Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants
Abstract
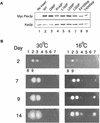
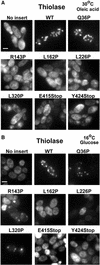
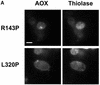
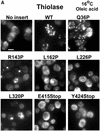
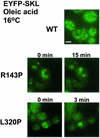
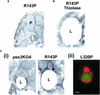
Pex3p is a peroxisomal integral membrane protein required early in peroxisome biogenesis, and Pex3p-deficient cells lack identifiable peroxisomes. Two temperature-sensitive pex3 mutant strains of the yeast Yarrowia lipolytica were made to investigate the role of Pex3p in the early stages of peroxisome biogenesis. In glucose medium at 16 degrees C, these mutants underwent de novo peroxisome biogenesis and exhibited early matrix protein sequestration into peroxisome-like structures found at the endoplasmic reticulum-rich periphery of cells or sometimes associated with nuclei. The de novo peroxisome biogenesis seemed unsynchronized, with peroxisomes occurring at different stages of development both within cells and between cells. Cells with peripheral nascent peroxisomes and cells with structures morphologically distinct from peroxisomes, such as semi/circular tubular structures that immunostained with antibodies to peroxisomal matrix proteins and to the endoplasmic reticulum-resident protein Kar2p, and that surrounded lipid droplets, were observed during up-regulation of peroxisome biogenesis in cells incubated in oleic acid medium at 16 degrees C. These structures were not detected in wild-type or Pex3p-deficient cells. Their role in peroxisome biogenesis remains unclear. Targeting of peroxisomal matrix proteins to these structures suggests that Pex3p directly or indirectly sequesters components of the peroxisome biogenesis machinery. Such a role is consistent with Pex3p overexpression producing cells with fewer, larger, and clustered peroxisomes.
Figures










References
-
- Aitchison JD, Szilard RK, Nuttley WM, Rachubinski RA. Antibodies directed against a yeast carboxyl-terminal peroxisomal targeting signal specifically recognize peroxisomal proteins from various yeasts. Yeast. 1992;8:721–734. - PubMed
-
- Ausubel FJ, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994.
-
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36:1211–1226. - PubMed
-
- Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11:R446–R449. - PubMed
-
- Chang CC, South S, Warren D, Jones J, Moser AB, Moser HW, Gould SJ. Metabolic control of peroxisome abundance. J Cell Sci. 1999;112:1579–1590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

