Global transcription analysis of Krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes
- PMID: 12631716
- PMCID: PMC151572
- DOI: 10.1091/mbc.e02-07-0422
Global transcription analysis of Krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes
Abstract
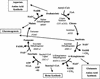
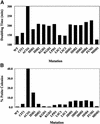
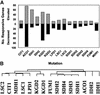
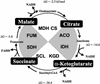
To understand the many roles of the Krebs tricarboxylic acid (TCA) cycle in cell function, we used DNA microarrays to examine gene expression in response to TCA cycle dysfunction. mRNA was analyzed from yeast strains harboring defects in each of 15 genes that encode subunits of the eight TCA cycle enzymes. The expression of >400 genes changed at least threefold in response to TCA cycle dysfunction. Many genes displayed a common response to TCA cycle dysfunction indicative of a shift away from oxidative metabolism. Another set of genes displayed a pairwise, alternating pattern of expression in response to contiguous TCA cycle enzyme defects: expression was elevated in aconitase and isocitrate dehydrogenase mutants, diminished in alpha-ketoglutarate dehydrogenase and succinyl-CoA ligase mutants, elevated again in succinate dehydrogenase and fumarase mutants, and diminished again in malate dehydrogenase and citrate synthase mutants. This pattern correlated with previously defined TCA cycle growth-enhancing mutations and suggested a novel metabolic signaling pathway monitoring TCA cycle function. Expression of hypoxic/anaerobic genes was elevated in alpha-ketoglutarate dehydrogenase mutants, whereas expression of oxidative genes was diminished, consistent with a heme signaling defect caused by inadequate levels of the heme precursor, succinyl-CoA. These studies have revealed extensive responses to changes in TCA cycle function and have uncovered new and unexpected metabolic networks that are wired into the TCA cycle.
Figures






References
-
- Anderson SL, Minard KI, McAlister-Henn L. Allosteric inhibition of NAD+-specific isocitrate dehydrogenase by a mitochondrial mRNA. Biochemistry. 2000;39:5623–5629. - PubMed
-
- Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. - PubMed
-
- Bojunga N, Entian K-D. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p) Mol Gen Genet. 1999;262:869–785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

