Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus
- PMID: 12631723
- PMCID: PMC151579
- DOI: 10.1091/mbc.e02-06-0317
Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus
Abstract
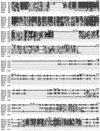
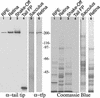
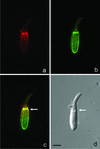
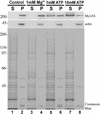
The striped bass has two retina-expressed class III myosin genes, each composed of a kinase, motor, and tail domain. We report the cloning, sequence analysis, and expression patterns of the long (Myo3A) and short (Myo3B) class III myosins, as well as cellular localization and biochemical characterization of the long isoform, Myo3A. Myo3A (209 kDa) is expressed in the retina, brain, testis, and sacculus, and Myo3B (155 kDa) is expressed in the retina, intestine, and testis. The tails of these two isoforms contain two highly conserved domains, 3THDI and 3THDII. Whereas Myo3B has three IQ motifs, Myo3A has nine IQ motifs, four in its neck and five in its tail domain. Myo3A localizes to actin filament bundles of photoreceptors and is concentrated in the calycal processes. An anti-Myo3A antibody decorates the actin cytoskeleton of rod inner/outer segments, and this labeling is reduced by the presence of ATP. The ATP-sensitive actin association is a feature characteristic of myosin motors. The numerous IQ motifs may play a structural or signaling role in the Myo3A, and its localization to calycal processes indicates that this myosin mediates a local function at this site in vertebrate photoreceptors.
Figures




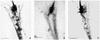

References
-
- Anderson DH, Johnson LV, Hageman GS. Vitronectin receptor expression and distribution at the photoreceptor-retinal pigment epithelial interface. J Comp Neurol. 1995;360:1–16. - PubMed
-
- Apweiler R, et al. Interpro—an integrated documentation resource for protein families, domains, and functional sites. Bioinformatics. 2000;16:1145–1150. - PubMed
-
- Arikawa K, Williams DS. Alpha-actinin and actin in the outer retina: a double immunoelectron microscopic study. Cell Motil Cytoskeleton. 1991;18:15–25. - PubMed
-
- Barnes JA, Gomes AV. PEST sequences in calmodulin-binding proteins. Mol Cell Biochem. 1995;149–150:17–27. - PubMed
-
- Barylko B, Binns DD, Albanesi JP. Regulation of the enzymatic and motor activities of myosin I. Biochim Biophys Acta. 2000;1496:23–35. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

