Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae
- PMID: 12631735
- PMCID: PMC151591
- DOI: 10.1091/mbc.e02-09-0609
Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae
Abstract
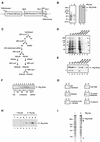
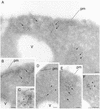
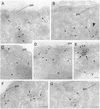
Tor1p and Tor2p kinases, targets of the immune-suppressive antibiotic rapamycin, are components of a highly conserved signaling network that couples nutrient availability and cell growth. To gain insight into the molecular basis underlying Tor-dependent signaling, we used cell fractionation and immunoaffinity chromatography to examine the physical environment of Tor2p. We found that the majority of Tor2p associates with a membrane-bound compartment along with at least four other proteins, Avo1p-Avo3p and Lst8p. Using immunogold electron microscopy, we observed that Tor2p, as well as Tor1p, localizes in punctate clusters to regions adjacent to the plasma membrane and within the cell interior, often in association with characteristic membranous tracks. Cell fractionation, coimmunoprecipitation, and immunogold electron microscopy experiments confirmed that Lst8 associates with both Tor2p as well as Tor1p at these membranous sites. In contrast, we find that Kog1, the yeast homologue of the mammalian Tor regulatory protein Raptor, interacts preferentially with Tor1p. These findings provide evidence for the existence of Tor signaling complexes that contain distinct as well as overlapping components. That these complexes colocalize to a membrane-bound compartment suggests an intimate relationship between membrane-mediated signaling and Tor activity.
Figures




References
-
- Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. - PubMed
-
- Andrade MA, Bork P. Heat repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. - PubMed
-
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of Ca2+-sensitive csg2Δ mutant. J Biol Chem. 1988;273:30688–30694. - PubMed
-
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan T-F, Zheng XFS. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

