ATLAS: a system to selectively identify human-specific L1 insertions
- PMID: 12632328
- PMCID: PMC1180347
- DOI: 10.1086/373939
ATLAS: a system to selectively identify human-specific L1 insertions
Abstract
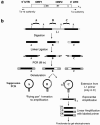
Retrotransposition of L1 LINEs (long interspersed elements) continues to sculpt the human genome. However, because recent insertions are dimorphic, they are not fully represented in sequence databases. Here, we have developed a system, termed "ATLAS" (amplification typing of L1 active subfamilies), that enables the selective amplification and display of DNA fragments containing the termini of human-specific L1s and their respective flanking sequences. We demonstrate that ATLAS is robust and that the resultant display patterns are highly reproducible, segregate in Centre d'Etude du Polymorphisme Humain pedigrees, and provide an individual-specific fingerprint. ATLAS also allows the identification of L1s that are absent from current genome databases, and we show that some of these L1s can retrotranspose at high frequencies in cultured human cells. Finally, we demonstrate that ATLAS also can identify single-nucleotide polymorphisms within a subset of older, primate-specific L1s. Thus, ATLAS provides a simple, high-throughput means to assess genetic variation associated with L1 retrotransposons.
Figures





References
Electronic-Database Information
-
- Authors' Web Site, http://www.le.ac.uk/ge/ajj/LINE1/ (for supplementary tables S1 and S2, as well as full-length L1 database and full-length L1 alignment)
-
- CEPH-Généthon Web Site, http://www.cephb.fr/ceph-genethon-map.html
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for rs2503417)
-
- Entrez Genome View, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi? (for NCBI Map Viewer)
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 - PubMed
-
- Alves G, Tatro A, Fanning T (1996) Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 176:39–44 - PubMed
-
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 - PubMed
-
- Buzdin A, Khodosevich K, Mamedov I, Vinogradova T, Lebedev Y, Hunsmann G, Sverdlov E (2002) A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics 79:413–422 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

