The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle
- PMID: 12634354
- PMCID: PMC150645
- DOI: 10.1128/jvi.77.7.3939-3949.2003
The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle
Abstract
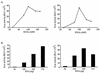
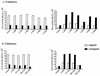
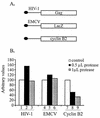
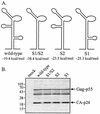
The 5' leader of the human immunodeficiency virus type 1 (HIV-1) genomic RNA contains highly structured domains involved in key steps of the viral life cycle. These RNA domains inhibit cap-dependent protein synthesis. Here we report that the HIV-1 5' leader harbors an internal ribosome entry site (IRES) capable of driving protein synthesis during the G(2)/M cell cycle phase in which cap-dependent initiation is inhibited. The HIV-1 IRES was delineated with bicistronic mRNAs in in vitro and ex vivo assays. The HIV-1 leader IRES spans nucleotides 104 to 336 and partially overlaps the major determinants of genomic RNA packaging. These data strongly suggest that, as for HIV-1 transcription, IRES-mediated translation initiation could play an important role in virus replication during virus-induced G(2)/M cell cycle arrest.
Figures




References
-
- Abbink, T. E., and B. Berkhout. A novel long-distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the gag start codon. J. Biol. Chem., in press. - PubMed
-
- Alizon, M., S. Wain-Hobson, L. Montagnier, and P. Sonigo. 1986. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell 46:63-74. - PubMed
-
- Attal, J., M. C. Theron, F. Taboit, M. Cajero-Juarez, G. Kann, P. Bolifraud, and L. M. Houdebine. 1996. The RU5 ('R') region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 392:220-224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

