The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations
- PMID: 12640112
- PMCID: PMC150720
- DOI: 10.1128/MCB.23.7.2264-2276.2003
The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations
Abstract
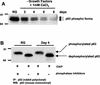
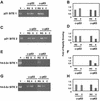
p63 is a recently identified homolog of p53 that is found in the basal layer of several stratified epithelial tissues such as the epidermis, oral mucosa, prostate, and urogenital tract. Studies with p63(-/-) mice and analysis of several human autosomal-dominant disorders with germ line p63 mutations suggest p63 involvement in maintaining epidermal stem cell populations. The p63 gene encodes six splice variants with reported transactivating or dominant-negative activities. The goals of the current study were to determine the splice variants that are expressed in primary human epidermal keratinocytes (HEKs) and the biochemical activity p63 has in these epithelial cell populations. We found that the predominant splice variant expressed in HEKs was Delta Np63 alpha, and it was present as a phosphorylated protein. During HEK differentiation, Delta Np63 alpha and p53 levels decreased, while expression of p53 target genes p21 and 14-3-3 sigma increased. Delta Np63 alpha had transcriptional repressor activity in vitro, and this activity was reduced in Delta Np63 alpha proteins containing point mutations, corresponding to those found in patients with Hay-Wells syndrome. Further, we show that Delta Np63 alpha and p53 can bind the p21 and 14-3-3 sigma promoters in vitro and in vivo, with decreased binding of p63 to these promoters during HEK differentiation. These data suggest that Delta Np63 alpha acts as a transcriptional repressor at select growth regulatory gene promoters in HEKs, and this repression likely plays an important role in the proliferative capacity of basal keratinocytes.
Figures





References
-
- Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. - PMC - PubMed
-
- Augustin, M., C. Bamberger, D. Paul, and H. Schmale. 1998. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homolog Ket to mouse chromosome 16. Mamm. Genome 9:899-902. - PubMed
-
- Celli, J., P. Duijf, B. Hamel, M. Bamshad, B. Kramer, A. Smits, R. Newbury-Ecob, R. Hennekam, G. Van Buggenhout, A. van Haeringen, C. Woods, A. van Essen, R. de Waal, G. Vriend, D. Haber, A. Yang, F. McKeon, H. Brunner, and H. van Bokhoven. 1999. Heterozygous germline muations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143-153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
