Phase variation of Ag43 is independent of the oxidation state of OxyR
- PMID: 12644490
- PMCID: PMC151510
- DOI: 10.1128/JB.185.7.2203-2209.2003
Phase variation of Ag43 is independent of the oxidation state of OxyR
Abstract
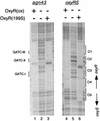
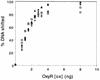
OxyR is a DNA binding protein that differentially regulates a cell's response to hydrogen peroxide-mediated oxidative stress. We previously reported that the reduced form of OxyR is sufficient for repression of transcription of agn43 from unmethylated template DNA, which is essential for deoxyadenosine methylase (Dam)- and OxyR-dependent phase variation of agn43. Here we provide evidence that the oxidized form of OxyR [OxyR(ox)] also represses agn43 transcription. In vivo, we found that exogenous addition of hydrogen peroxide, sufficient to oxidize OxyR, did not affect the expression of agn43. OxyR(ox) repressed in vitro transcription but only from an unmethylated agn43 template. The -10 sequence of the promoter and three Dam target sequences were protected in an in vitro DNase I footprint assay by OxyR(ox). Furthermore, OxyR(ox) bound to the agn43 regulatory region DNA with an affinity similar to that for the regulatory regions of katG and oxyS, which are activated by OxyR(ox), indicating that binding at agn43 can occur at biologically relevant concentrations. OxyR-dependent regulation of Ag43 expression is therefore unusual in firstly that OxyR binding at agn43 is dependent on the methylation state of Dam target sequences in its binding site and secondly that OxyR-dependent repression appears to be independent of hydrogen-peroxide mediated oxidative stress and the oxidation state of OxyR.
Figures


Similar articles
-
Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation.J Bacteriol. 2002 Jun;184(12):3338-47. doi: 10.1128/JB.184.12.3338-3347.2002. J Bacteriol. 2002. PMID: 12029051 Free PMC article.
-
Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant.Mol Microbiol. 2002 Apr;44(2):521-32. doi: 10.1046/j.1365-2958.2002.02918.x. Mol Microbiol. 2002. PMID: 11972788
-
Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription.Mol Microbiol. 2000 Feb;35(4):877-87. doi: 10.1046/j.1365-2958.2000.01762.x. Mol Microbiol. 2000. PMID: 10692164
-
The OxyR regulon.Antonie Van Leeuwenhoek. 1990 Oct;58(3):157-61. doi: 10.1007/BF00548927. Antonie Van Leeuwenhoek. 1990. PMID: 2256675 Review.
-
Phase-variable outer membrane proteins in Escherichia coli.FEMS Immunol Med Microbiol. 1996 Dec 1;16(2):63-76. doi: 10.1111/j.1574-695X.1996.tb00124.x. FEMS Immunol Med Microbiol. 1996. PMID: 8988388 Review.
Cited by
-
Establishing and maintaining sequestration of Dam target sites for phase variation of agn43 in Escherichia coli.J Bacteriol. 2010 Apr;192(7):1937-45. doi: 10.1128/JB.01629-09. Epub 2010 Jan 29. J Bacteriol. 2010. PMID: 20118257 Free PMC article.
-
Control of gene expression at a bacterial leader RNA, the agn43 gene encoding outer membrane protein Ag43 of Escherichia coli.J Bacteriol. 2014 Aug;196(15):2728-35. doi: 10.1128/JB.01680-14. Epub 2014 May 16. J Bacteriol. 2014. PMID: 24837285 Free PMC article.
-
OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis.FEMS Microbiol Lett. 2008 May;282(2):188-95. doi: 10.1111/j.1574-6968.2008.01116.x. Epub 2008 Mar 18. FEMS Microbiol Lett. 2008. PMID: 18355277 Free PMC article.
-
Diarrhea-associated biofilm formed by enteroaggregative Escherichia coli and aggregative Citrobacter freundii: a consortium mediated by putative F pili.BMC Microbiol. 2010 Feb 23;10:57. doi: 10.1186/1471-2180-10-57. BMC Microbiol. 2010. PMID: 20175929 Free PMC article.
-
DNA Methylation.EcoSal Plus. 2014 May;6(1):10.1128/ecosalplus.ESP-0003-2013. doi: 10.1128/ecosalplus.ESP-0003-2013. EcoSal Plus. 2014. PMID: 26442938 Free PMC article.
References
-
- Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
-
- Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

