Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B(1)4
- PMID: 12644492
- PMCID: PMC151492
- DOI: 10.1128/JB.185.7.2219-2226.2003
Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B(1)4
Abstract
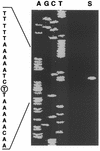
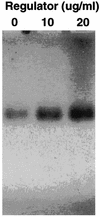
The xylanase gene cluster from the rumen anaerobe Prevotella bryantii B(1)4 was found to include a gene (xynR) that encodes a multidomain regulatory protein and is downstream from the xylanase and beta-xylosidase genes xynA and xynB. Additional genes identified upstream of xynA and xynB include xynD, which encodes an integral membrane protein that has homology with Na:solute symporters; xynE, which is related to the genes encoding acylhydrolases and arylesterases; and xynF, which has homology with the genes encoding alpha-glucuronidases. XynR includes, in a single 833-amino-acid polypeptide, a putative input domain unrelated to other database sequences, a likely transmembrane domain, histidine kinase motifs, response regulator sequences, and a C-terminal AraC-type helix-turn-helix DNA binding domain. Two transcripts (3.7 and 5.8 kb) were detected with a xynA probe, and the start site of the 3.7-kb transcript encoding xynABD was mapped to a position upstream of xynD. The DNA binding domain of XynR was purified after amplification and overexpression in Escherichia coli and was found to bind to a 141-bp DNA fragment from the region immediately upstream of xynD. In vitro transcription assays demonstrated that XynR stimulates transcription of the 3.7-kb transcript. We concluded that XynR acts as a positive regulator that activates expression of xynABD in P. bryantii B(1)4. This is the first regulatory protein that demonstrates significant homology with the two-component regulatory protein superfamily and has been shown to be involved in the regulation of polysaccharidase gene expression.
Figures
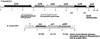





References
-
- Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix DNA-binding motif. J. Biol. Chem. 264:1903-1906. - PubMed
-
- Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. - PubMed
-
- Dehority, B. A. 1991. Effects of microbial synergism on fiber digestion in the rumen. Proc. Nutr. Soc. 50:149-159. - PubMed
-
- Flint, H. J., and C. S. Stewart. 1999. Bacteroides and Prevotella, p. 198-202. In R. K. Robinson, C. A. Batt, and P. Patel (ed.), Encyclopedia of food microbiology. Academic Press, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

