Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization
- PMID: 12644654
- PMCID: PMC166867
- DOI: 10.1104/pp.102.016436
Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization
Abstract
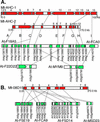
Arabidopsis and Medicago truncatula represent sister clades within the dicot subclass Rosidae. We used genetic map-based and bacterial artificial chromosome sequence-based approaches to estimate the level of synteny between the genomes of these model plant species. Mapping of 82 tentative orthologous gene pairs reveals a lack of extended macrosynteny between the two genomes, although marker collinearity is frequently observed over small genetic intervals. Divergence estimates based on non-synonymous nucleotide substitutions suggest that a majority of the genes under analysis have experienced duplication in Arabidopsis subsequent to divergence of the two genomes, potentially confounding synteny analysis. Moreover, in cases of localized synteny, genetically linked loci in M. truncatula often share multiple points of synteny with Arabidopsis; this latter observation is consistent with the large number of segmental duplications that compose the Arabidopsis genome. More detailed analysis, based on complete sequencing and annotation of three M. truncatula bacterial artificial chromosome contigs suggests that the two genomes are related by networks of microsynteny that are often highly degenerate. In some cases, the erosion of microsynteny could be ascribed to the selective gene loss from duplicated loci, whereas in other cases, it is due to the absence of close homologs of M. truncatula genes in Arabidopsis.
Figures



References
-
- Acarkan A, Rossberg M, Koch M, Schmidt R. Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J. 2000;23:55–62. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Bancroft I. Duplicate and diverge: the evolution of plant genome microstructure. Trends Genet. 2001;17:89–93. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

