Genetic diversity of Eurycoma longifolia inferred from single nucleotide polymorphisms
- PMID: 12644679
- PMCID: PMC166889
- DOI: 10.1104/pp.012492
Genetic diversity of Eurycoma longifolia inferred from single nucleotide polymorphisms
Abstract
Eurycoma longifolia Jack. is a treelet that grows in the forests of Southeast Asia and is widely used throughout the region because of its reported medicinal properties. Widespread harvesting of wild-grown trees has led to rapid thinning of natural populations, causing a potential decrease in genetic diversity among E. longifolia. Suitable genetic markers would be very useful for propagation and breeding programs to support conservation of this species, although no such markers currently exist. To meet this need, we have applied a genome complexity reduction strategy to identify a series of single nucleotide polymorphisms (SNPs) within the genomes of several E. longifolia accessions. We have found that the occurrence of these SNPs reflects the geographic origins of individual plants and can distinguish different natural populations. This work demonstrates the rapid development of molecular genetic markers in species for which little or no genomic sequence information is available. The SNP markers that we have developed in this study will also be useful for identifying genetic fingerprints that correlate with other properties of E. longifolia, such as high regenerability or the appearance of bioactive metabolites.
Figures

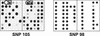
References
-
- Chan KL, O'Neill MJ, Phillipson JD, Warhurst DC. Plants as source of antimalarial drugs: Part 3. Eurycoma longifolia. Planta Med Apr. 1986;2:105–107. - PubMed
-
- Ching A, Rafalski A. Rapid genetic mapping of ESTs using SNP pyrosequencing and indel analysis. Cell Mol Biol Lett. 2002;7:803–810. - PubMed
-
- Creswell A, Sackville Hamilton NR, Roy AK, Viegas BMF. Use of amplified fragment length polymorphism markers to assess genetic diversity of Lolium sp. from Portugal. Mol Ecol. 2001;10:229–241. - PubMed
-
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 1987;19:11–15.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

