Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade
- PMID: 12644681
- PMCID: PMC166891
- DOI: 10.1104/pp.102.014928
Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade
Abstract
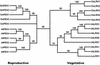
We have searched the Arabidopsis and rice (Oryza sativa) genomes for homologs of LRX1, an Arabidopsis gene encoding a novel type of cell wall protein containing a leucine-rich repeat (LRR) and an extensin domain. Eleven and eight LRX (LRR/EXTENSIN) genes have been identified in these two plant species, respectively. The LRX gene family encodes proteins characterized by a short N-terminal domain, a domain with 10 LRRs, a cysteine-rich motif, and a variable C-terminal extensin-like domain. Phylogenetic analysis performed on the conserved domains indicates the existence of two major clades of LRX proteins that arose before the eudicot/monocot divergence and then diversified independently in each lineage. In Arabidopsis, gene expression studies by northern hybridization and promoter::uidA fusions showed that the two phylogenetic clades represent a specialization into "reproductive" and "vegetative" LRXs. The four Arabidopsis genes of the "reproductive" clade are specifically expressed in pollen, whereas the seven "vegetative" genes are predominantly expressed in various sporophytic tissues. This separation into two expression classes is also supported by previous studies on maize (Zea mays) and tomato (Lycopersicon esculentum) LRX homologs and by information on available rice ESTs. The strong conservation of the amino acids responsible for the putative recognition specificity of the LRR domain throughout the family suggests that the LRX proteins interact with similar ligands.
Figures




References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R, Brutlog D, Karp P, Lathrop R, Searls D, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: American Association for Artificial Intelligence Press; 1994. pp. 28–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

