Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases
- PMID: 12646273
- PMCID: PMC1351391
- DOI: 10.1016/s0003-9861(03)00012-2
Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases
Abstract
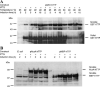
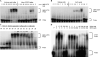
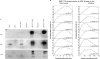
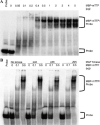
Tristetraprolin (TTP) is an mRNA-binding protein, but studies of this interaction have been difficult due to problems with the purification of recombinant TTP. In the present study, we expressed human and mouse TTP as glutathione S-transferase and maltose-binding protein (MBP) fusion proteins in Escherichia coli, and purified them by affinity resins and Mono Q chromatography. TTP cleaved from the fusion protein was identified by immunoblotting, MALDI-MS, and protein sequencing, and was further purified to homogeneity by continuous-elution SDS-gel electrophoresis. Purified recombinant TTP bound to the AU-rich element of tumor necrosis factor-alpha (TNFalpha) mRNA and this binding was dependent on Zn(2+). Results from sizing columns suggested that the active species might be in the form of an oligomer of MBP-TTP. Recombinant TTP was phosphorylated by three members of the mitogen-activated protein (MAP) kinase family, p42, p38, and JNK, with half-maximal phosphorylation occurring at approximately 0.5, 0.25, and 0.25 microM protein, respectively. Phosphorylation by these kinases did not appear to affect the ability of TTP to bind to TNFalpha mRNA under the assay conditions. This study describes a procedure for purifying nonfusion protein TTP to homogeneity, demonstrates that TTP's RNA-binding activity is zinc dependent, and that TTP can be phosphorylated by JNK as well as by the other members of the greater MAP kinase family.
Figures





Similar articles
-
Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin.Expert Rev Proteomics. 2007 Dec;4(6):711-26. doi: 10.1586/14789450.4.6.711. Expert Rev Proteomics. 2007. PMID: 18067411 Free PMC article. Review.
-
Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications.Biochemistry. 2004 Nov 2;43(43):13724-38. doi: 10.1021/bi049014y. Biochemistry. 2004. PMID: 15504035 Free PMC article.
-
MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding.J Biol Chem. 2004 Mar 12;279(11):10176-84. doi: 10.1074/jbc.M310486200. Epub 2003 Dec 19. J Biol Chem. 2004. PMID: 14688255
-
Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability.Mol Cell Biol. 2001 Oct;21(19):6461-9. doi: 10.1128/MCB.21.9.6461-6469.2001. Mol Cell Biol. 2001. PMID: 11533235 Free PMC article.
-
AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors.Biochem Soc Trans. 2002 Nov;30(Pt 6):952-8. doi: 10.1042/bst0300952. Biochem Soc Trans. 2002. PMID: 12440953 Review.
Cited by
-
Tumor-intrinsic CDC42BPB confers resistance to anti-PD-1 immune checkpoint blockade in breast cancer.Mol Ther. 2024 Oct 2;32(10):3669-3682. doi: 10.1016/j.ymthe.2024.07.021. Epub 2024 Jul 31. Mol Ther. 2024. PMID: 39086134
-
Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin.Expert Rev Proteomics. 2007 Dec;4(6):711-26. doi: 10.1586/14789450.4.6.711. Expert Rev Proteomics. 2007. PMID: 18067411 Free PMC article. Review.
-
The requirement of c-Jun N-terminal kinase 2 in regulation of hypoxia-inducing factor-1α mRNA stability.J Biol Chem. 2012 Oct 5;287(41):34361-71. doi: 10.1074/jbc.M112.365882. Epub 2012 Aug 21. J Biol Chem. 2012. PMID: 22910906 Free PMC article.
-
C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival.Genes Dev. 2006 Aug 15;20(16):2279-92. doi: 10.1101/gad.384506. Genes Dev. 2006. PMID: 16912277 Free PMC article.
-
Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways.Mol Cell Biol. 2006 Mar;26(6):2408-18. doi: 10.1128/MCB.26.6.2408-2418.2006. Mol Cell Biol. 2006. PMID: 16508015 Free PMC article.
References
-
- DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. J. Biol. Chem. 1990;265:19185–19191. - PubMed
-
- Lai WS, Stumpo DJ, Blackshear PJ. J. Biol. Chem. 1990;265:16556–16563. - PubMed
-
- Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. J. Biol. Chem. 2000;275:17827–17837. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Science. 1998;281:1001–1005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

