Designing isoform-specific peptide disruptors of protein kinase A localization
- PMID: 12646696
- PMCID: PMC153050
- DOI: 10.1073/pnas.2628038100
Designing isoform-specific peptide disruptors of protein kinase A localization
Abstract
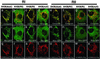
A kinase-anchoring proteins (AKAPs) coordinate cAMP-mediated signaling by binding and localizing cAMP-dependent protein kinase (PKA), using an amphipathic helical docking motif. Peptide disruptors of PKA localization that mimic this helix have been used successfully to assess the involvement of PKA in specific signaling pathways. However, these peptides were developed as disruptors for the type II regulatory subunit (RII) even though both RI and RII isoforms can bind to AKAPs and have discrete functions. To evaluate the effects of each localized isoform, we designed peptides that specifically bind to either RI or RII. Using a peptide array, we have defined the minimal binding sequence of dual specific-AKAP 2 (d-AKAP2), which binds tightly to both RI and RII. Side-chain requirements for affinity and isoform specificity were evaluated by using a peptide substitution array where each position along the A kinase binding domain of d-AKAP2 was substituted by the other 19 l-amino acids. This array comprises 513 single-site substitution analogs of the d-AKAP2 sequence. Peptides containing single and multiple mutations were evaluated in a quantitative fluorescence binding assay and a cell-based colocalization assay. This strategy has allowed us to design peptides with high affinity (K(D) = 1-2 nM) and high specificity for RIalpha versus RIIalpha. These isoform-specific peptides will be invaluable tools to evaluate functional differences between localized RI and RII PKA and are RIalpha-specific disruptors. This array-based analysis also provides a foundation for biophysical analysis of this docking motif.
Figures
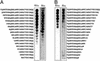

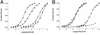
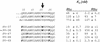
References
-
- Taylor S S, Buechler J A, Yonemoto W. Annu Rev Biochem. 1990;59:971–1005. - PubMed
-
- Colledge M, Scott J D. Trends Cell Biol. 1999;9:216–221. - PubMed
-
- Doskeland S O, Maronde E, Gjertsen B T. Biochim Biophys Acta. 1993;1178:249–258. - PubMed
-
- Feliciello A, Gottesman M E, Avvedimento E V. J Mol Biol. 2001;308:99–114. - PubMed
-
- Skalhegg B S, Tasken K. Front Biosci. 2000;5:D678–D693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

