Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster
- PMID: 12646701
- PMCID: PMC153048
- DOI: 10.1073/pnas.0638023100
Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster
Abstract
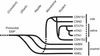
Gene duplication creates evolutionary novelties by using older tools in new ways. We have identified evidence that the genes for enamel matrix proteins (EMPs), milk caseins, and salivary proteins comprise a family descended from a common ancestor by tandem gene duplication. These genes remain linked, except for one EMP gene, amelogenin. These genes show common structural features and are expressed in ontogenetically similar tissues. Many of these genes encode secretory Ca-binding phosphoproteins, which regulate the Ca-phosphate concentration of the extracellular environment. By exploiting this fundamental property, these genes have subsequently diversified to serve specialized adaptive functions. Casein makes milk supersaturated with Ca-phosphate, which was critical to the successive mammalian divergence. The innovation of enamel led to mineralized feeding apparatus, which enabled active predation of early vertebrates. The EMP genes comprise a subfamily not identified previously. A set of genes for dentine and bone extracellular matrix proteins constitutes an additional cluster distal to the EMP gene cluster, with similar structural features to EMP genes. The duplication and diversification of the primordial genes for enameldentinebone extracellular matrix may have been important in core vertebrate feeding adaptations, the mineralized skeleton, the evolution of saliva, and, eventually, lactation. The order of duplication events may help delineate early events in mineralized skeletal formation, which is a major characteristic of vertebrates.
Figures
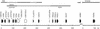


References
-
- Ohno S. Evolution by Gene Duplication. New York: Springer; 1970.
-
- Zeichner-David M, Diekwisch T, Fincham A, Lau E, MacDougall M, Moradian-Oldak J, Simmer J, Snead M, Slavkin H C. Int J Dev Biol. 1995;39:69–92. - PubMed
-
- Thesleff I, Sharpe P. Mech Dev. 1997;67:111–123. - PubMed
-
- Fincham A G, Moradian-Oldak J, Simmer J P. J Struct Biol. 1999;126:270–299. - PubMed
-
- Ginger M R, Grigor M R. Comp Biochem Physiol B Biochem Mol Biol. 1999;124:133–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

