Noncovalent scFv multimers of tumor-targeting anti-Lewis(y) hu3S193 humanized antibody
- PMID: 12649432
- PMCID: PMC2323837
- DOI: 10.1110/ps.0228503
Noncovalent scFv multimers of tumor-targeting anti-Lewis(y) hu3S193 humanized antibody
Abstract
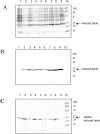
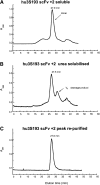
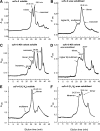
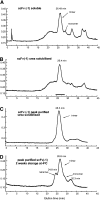
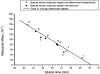
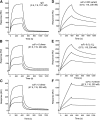
Single-chain variable fragments (scFvs) of anti-Lewis(y) hu3S193 humanized antibody were constructed by joining the V(H) and V(L) domains with either +2 residues, +1 residue, or by directly linking the domains. In addition two constructs were synthesized in which one or two C-terminal residues of the V(H) domain were removed (-1 residue, -2 residue) and then joined directly to the V(L) domain. An scFv construct in the reverse orientation with the V(L) joined directly to the V(H) domain was also synthesized. Upon transformation into Escherichia coli all scFv constructs expressed active protein. Binding activity, multimeric status, and multivalent properties were assessed by flow cytometry, size exclusion chromatography, and biosensor analysis. The results for hu3S193 scFvs are consistent with the paradigm that scFvs with a linker of +3 residues or more associate to form a non-covalent dimer, and those with a shorter linker or directly linked associate predominantly to form a non-covalent trimer and tetramer that are in equilibrium. While the association of V domains to form either a dimer or trimer/tetramer is governed by the length of the linker, the stability of the trimer/tetramer in the equilibrium mixture is dependent on the affinity of the interaction of the individual V domains to associate to form the larger Fv module.
Figures

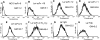

Similar articles
-
Tumor targeting by a multivalent single-chain Fv (scFv) anti-Lewis Y antibody construct.Cancer Biother Radiopharm. 2008 Aug;23(4):411-23. doi: 10.1089/cbr.2007.0450. Cancer Biother Radiopharm. 2008. PMID: 18771345 Free PMC article.
-
ScFv multimers of the anti-neuraminidase antibody NC10: shortening of the linker in single-chain Fv fragment assembled in V(L) to V(H) orientation drives the formation of dimers, trimers, tetramers and higher molecular mass multimers.Protein Eng. 2000 Aug;13(8):565-74. doi: 10.1093/protein/13.8.565. Protein Eng. 2000. PMID: 10964986
-
Single-chain Fv multimers of the anti-neuraminidase antibody NC10: the residue at position 15 in the V(L) domain of the scFv-0 (V(L)-V(H)) molecule is primarily responsible for formation of a tetramer-trimer equilibrium.Protein Eng. 2003 Jan;16(1):47-56. doi: 10.1093/proeng/gzg006. Protein Eng. 2003. PMID: 12646692
-
High avidity scFv multimers; diabodies and triabodies.J Immunol Methods. 1999 Dec 10;231(1-2):177-89. doi: 10.1016/s0022-1759(99)00157-x. J Immunol Methods. 1999. PMID: 10648937 Review.
-
Dimeric and trimeric antibodies: high avidity scFvs for cancer targeting.Biomol Eng. 2001 Oct 15;18(3):95-108. doi: 10.1016/s1389-0344(01)00090-9. Biomol Eng. 2001. PMID: 11566601 Review.
Cited by
-
Cancer therapeutic trispecific antibodies recruiting both T and natural killer cells to cancer cells.Oncol Rep. 2023 Dec;50(6):212. doi: 10.3892/or.2023.8649. Epub 2023 Oct 20. Oncol Rep. 2023. PMID: 37859608 Free PMC article.
-
Optimization of the crystallizability of a single-chain antibody fragment.Acta Crystallogr F Struct Biol Commun. 2014 Dec 1;70(Pt 12):1701-6. doi: 10.1107/S2053230X1402247X. Epub 2014 Nov 14. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25484230 Free PMC article.
-
Tumor targeting by a multivalent single-chain Fv (scFv) anti-Lewis Y antibody construct.Cancer Biother Radiopharm. 2008 Aug;23(4):411-23. doi: 10.1089/cbr.2007.0450. Cancer Biother Radiopharm. 2008. PMID: 18771345 Free PMC article.
-
A general chemical synthesis platform for crosslinking multivalent single chain variable fragments.Org Biomol Chem. 2012 Feb 28;10(8):1521-6. doi: 10.1039/c0ob01259a. Epub 2011 Dec 1. Org Biomol Chem. 2012. PMID: 22132412 Free PMC article.
-
Sortase-catalyzed in vitro functionalization of a HER2-specific recombinant Fab for tumor targeting of the plant cytotoxin gelonin.MAbs. 2014 Mar-Apr;6(2):354-66. doi: 10.4161/mabs.27444. Epub 2013 Dec 9. MAbs. 2014. PMID: 24492291 Free PMC article.
References
-
- Adams, G.P. and Schier, R. 1999. Generating improved single-chain Fv molecules for tumor targeting. J. Immunol. Methods 231 249–260. - PubMed
-
- Adams, G.P., Schier, R., Marshall, K., Wolf, E.J., McCall, A.M., Marks, J.D., and Weiner, L.M. 1998b. Increased affinity leads to improved selective tumor delivery of single-chain Fv antibodies. Cancer Res. 58 485–490. - PubMed
-
- Adams, G.P., Shaller, C.C., Chappell, L.L., Wu, C., Horak, E.M., Simmons, H.H., Litwin, S., Marks, J.D., Weiner, L.M., and Brechbiel, M.W. 2000. Delivery of the α-emitting radioisotope bismuth-213 to solid tumors via single-chain Fv and diabody molecules. Nucl. Med. Biol. 27 339–346. - PubMed
-
- Atwell, J.L., Breheney, K.A., Lawrence, L.J., McCoy, A.J., Kortt, A.A., and Hudson, P.J. 1999. scFv multimers of the anti-neuraminidase antibody NC10: Length of the linker between VH and VL domains dictates precisely the transition between diabodies and triabodies. Protein Eng. 12 597–604. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

