Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA
- PMID: 12649489
- PMCID: PMC1370404
- DOI: 10.1261/rna.2200903
Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA
Abstract
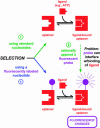
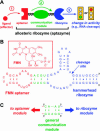
Switches and sensors play important roles in our everyday lives. The chemical properties of RNA make it amenable for use as a switch or sensor, both artificially and in nature. This review focuses on recent advances in artificial RNA switches and sensors. Researchers have been applying classical biochemical principles such as allostery in elegant ways that are influencing the development of biosensors and other applications. Particular attention is given here to allosteric ribozymes (aptazymes) that are regulated by small organic molecules, by proteins, or by oligonucleotides. Also discussed are ribozymes whose activities are controlled by various nonallosteric strategies.
Figures

References
-
- Araki, M., Hashima, M., Okuno, Y., and Sugiura, Y. 2001. Coupling between substrate binding and allosteric regulation in ribozyme catalysis. Bioorg. Med. Chem. 9: 1155–1163. - PubMed
-
- Breaker, R.R. 2002. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotech. 13: 31–39. - PubMed
-
- Burke, D.H., Ozerova, N.D., and Nilsen-Hamilton, M. 2002. Allosteric hammerhead ribozyme TRAPs. Biochemistry 41: 6588–6594. - PubMed
-
- Famulok, M. and Verma, S. 2002. In vivo-applied functional RNAs as tools in proteomics and genomics research. Trends Biotechnol. 20: 462–466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
