The relationship between hypomethylation and CpG island methylation in colorectal neoplasia
- PMID: 12651628
- PMCID: PMC1851239
- DOI: 10.1016/S0002-9440(10)63932-6
The relationship between hypomethylation and CpG island methylation in colorectal neoplasia
Abstract
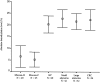
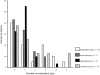
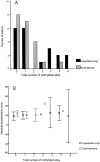
Tumors are often characterized by an imbalance in cytosine methylation as manifested both by hypermethylation of CpG islands and by genome hypomethylation. These epigenetic changes were assessed in colorectal neoplasia to determine whether they arose through a common mechanism or indeed were distinct and unrelated phenomena. Fresh representative samples of adenomas, hyperplastic polyps, colorectal cancers, and normal mucosa were used in this study. Global methylation levels were measured by analyzing the methyl-accepting capacity of DNA. Methylation of p16, hMLH1, and MINT 1, 2, 12, and 31 were assessed by bisulfite polymerase chain reaction. Microsatellite status was determined by polymerase chain reaction using six markers and hMLH1 and proliferating cell nuclear antigen expression was assessed by immunohistochemistry. Normal colonic mucosa had a higher endogenous 5-methyl cytosine content than all proliferative lesions of the colon (P < 0.001). The extent of demethylation in hyperplastic polyps and adenomas was significantly related to its proliferative rate. Right-sided hyperplastic polyps were more likely to be methylated than adenomas (odds ratio, 2.3; confidence interval, 1.1 to 4.6). There was no relationship between the level of global hypomethylation and hypermethylation. Some hyperplastic colorectal polyps have a propensity to develop dense CpG island methylation. Hypermethylation and hypomethylation contribute separately to the process of carcinogenesis.
Figures


Comment in
-
Aberrant DNA methylation: have we entered the era of more than one type of colorectal cancer?Am J Pathol. 2003 Apr;162(4):1043-5. doi: 10.1016/S0002-9440(10)63900-4. Am J Pathol. 2003. PMID: 12651596 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

