Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase
- PMID: 12651944
- PMCID: PMC153055
- DOI: 10.1073/pnas.0730835100
Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase
Abstract
The expression of activation-induced cytidine deaminase (AID) is prerequisite to a "trifecta" of key molecular events in B cells: class-switch recombination and somatic hypermutation in humans and mice and gene conversion in chickens. Although this critically important enzyme shares common sequence motifs with apolipoprotein B mRNA-editing enzyme, and exhibits deaminase activity on free deoxycytidine in solution, it has not been shown to act on either RNA or DNA. Recent mutagenesis data in Escherichia coli suggest that AID may deaminate dC on DNA, but its putative biochemical activities on either DNA or RNA remained a mystery. Here, we show that AID catalyzes deamination of dC residues on single-stranded DNA in vitro but not on double-stranded DNA, RNA-DNA hybrids, or RNA. Remarkably, it has no measurable deaminase activity on single-stranded DNA unless pretreated with RNase to remove inhibitory RNA bound to AID. AID catalyzes dC --> dU deamination activity most avidly on double-stranded DNA substrates containing a small "transcription-like" single-stranded DNA bubble, suggesting a targeting mechanism for this enigmatic enzyme during somatic hypermutation.
Figures
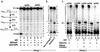


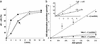
References
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. - PubMed
-
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. - PubMed
-
- Martin A, Bardwell P D, Woo C J, Fan M, Shulman M J, Scharff M D. Nature. 2002;415:802–806. - PubMed
-
- Yoshikawa K, Okazaki I M, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Science. 2002;296:2033–2036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

