Major histocompatibility complex heterozygote superiority during coinfection
- PMID: 12654829
- PMCID: PMC152037
- DOI: 10.1128/IAI.71.4.2079-2086.2003
Major histocompatibility complex heterozygote superiority during coinfection
Abstract
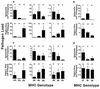
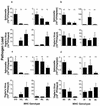
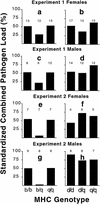
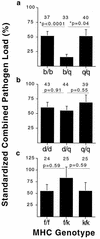
Genes of the major histocompatibility complex (MHC) play a critical role in immune recognition, and many alleles confer susceptibility to infectious and autoimmune diseases. How these deleterious alleles persist in populations is controversial. One hypothesis postulates that MHC heterozygote superiority emerges over multiple infections because MHC-mediated resistance is generally dominant and many allele-specific susceptibilities to pathogens will be masked by the resistant allele in heterozygotes. We tested this hypothesis by using experimental coinfections with Salmonella enterica (serovar Typhimurium C5TS) and Theiler's murine encephalomyelitis virus (TMEV) in MHC-congenic mouse strains where one haplotype was resistant to Salmonella and the other was resistant to TMEV. MHC heterozygotes were superior to both homozygotes in 7 out of 8 comparisons (P = 0.0024), and the mean standardized pathogen load of heterozygotes was reduced by 41% over that of homozygotes (P = 0.01). In contrast, no heterozygote superiority was observed when the MHC haplotype combinations had similar susceptibility profiles to the two pathogens. This is the first experimental evidence for MHC heterozygote superiority against multiple pathogens, a mechanism that would contribute to the evolution of MHC diversity and explain the persistence of alleles conferring susceptibility to disease.
Figures
Similar articles
-
Infection-dependent phenotypes in MHC-congenic mice are not due to MHC: can we trust congenic animals?BMC Immunol. 2004 Jul 9;5:14. doi: 10.1186/1471-2172-5-14. BMC Immunol. 2004. PMID: 15245582 Free PMC article.
-
MHC heterozygosity confers a selective advantage against multiple-strain infections.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11260-4. doi: 10.1073/pnas.162006499. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177415 Free PMC article.
-
Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice.Genetics. 2007 Aug;176(4):2501-8. doi: 10.1534/genetics.107.074815. Epub 2007 Jul 1. Genetics. 2007. PMID: 17603099 Free PMC article.
-
The genetics of the persistent infection and demyelinating disease caused by Theiler's virus.Annu Rev Microbiol. 2005;59:279-98. doi: 10.1146/annurev.micro.59.030804.121242. Annu Rev Microbiol. 2005. PMID: 16153171 Review.
-
Two models of multiple sclerosis: experimental allergic encephalomyelitis (EAE) and Theiler's murine encephalomyelitis virus (TMEV) infection. A pathological and immunological comparison.Microsc Res Tech. 1995 Oct 15;32(3):215-29. doi: 10.1002/jemt.1070320305. Microsc Res Tech. 1995. PMID: 8527856 Free PMC article. Review.
Cited by
-
Major histocompatibility complex class II DAB alleles associated with intestinal parasite load in the vulnerable Chinese egret (Egretta eulophotes).Ecol Evol. 2016 Jun 7;6(13):4421-34. doi: 10.1002/ece3.2226. eCollection 2016 Jul. Ecol Evol. 2016. PMID: 27386085 Free PMC article.
-
Marriage does not relate to major histocompatibility complex: a genetic analysis based on 3691 couples.Proc Biol Sci. 2020 Oct 14;287(1936):20201800. doi: 10.1098/rspb.2020.1800. Epub 2020 Oct 7. Proc Biol Sci. 2020. PMID: 33023409 Free PMC article.
-
Major histocompatibility complex controls the trajectory but not host-specific adaptation during virulence evolution of the pathogenic fungus Cryptococcus neoformans.Proc Biol Sci. 2004 Aug 7;271(1548):1557-64. doi: 10.1098/rspb.2004.2736. Proc Biol Sci. 2004. PMID: 15306300 Free PMC article.
-
Major histocompatibility complex based resistance to a common bacterial pathogen of amphibians.PLoS One. 2008 Jul 16;3(7):e2692. doi: 10.1371/journal.pone.0002692. PLoS One. 2008. PMID: 18629002 Free PMC article.
-
Signatures of historical demography and pathogen richness on MHC class I genes.Immunogenetics. 2012 Mar;64(3):165-75. doi: 10.1007/s00251-011-0576-y. Epub 2011 Sep 23. Immunogenetics. 2012. PMID: 21947542
References
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Apanius, V., D. Penn, P. Slev, L. R. Ruff, and W. K. Potts. 1997. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 17:179-224. - PubMed
-
- Baum, H., and N. A. Staines. 1997. MHC-derived peptides and the CD4+ T-cell repertoire: implications for autoimmune disease. Cytokines Cell. Mol. Ther. 3:115-125. - PubMed
-
- Behnke, J. M., and F. N. Wahid. 1991. Immunological relationships during primary infection with Heligmosomoides polygyrus (Nematospiroides dubius): H-2 linked genes determine worm survival. Parasitology 103:157-164. - PubMed
-
- Bertoletti, A., A. Sette, F. V. Chisari, A. Penna, M. Levrero, M. De Carli, F. Fiaccadori, and C. Ferrari. 1994. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature 369:407-410. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

