Molecular recognition properties of IGS-mediated reactions catalyzed by a Pneumocystis carinii group I intron
- PMID: 12655009
- PMCID: PMC152796
- DOI: 10.1093/nar/gkg280
Molecular recognition properties of IGS-mediated reactions catalyzed by a Pneumocystis carinii group I intron
Abstract
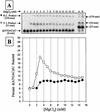
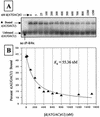
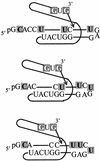
We report the development, analysis and use of a new combinatorial approach to analyze the substrate sequence dependence of the suicide inhibition, cyclization, and reverse cyclization reactions catalyzed by a group I intron from the opportunistic pathogen Pneumocystis carinii. We demonstrate that the sequence specificity of these Internal Guide Sequence (IGS)-mediated reactions is not high. In addition, the sequence specificity of suicide inhibition decreases with increasing MgCl(2) concentration, reverse cyclization is substantially more sequence specific than suicide inhibition, and multiple reverse cyclization products occur, in part due to the formation of multiple cyclization intermediates. Thermodynamic analysis reveals that a base pair at position -4 of the resultant 5' exon-IGS (P1) helix is crucial for tertiary docking of the P1 helix into the catalytic core of the ribozyme in the suicide inhibition reaction. In contrast to results reported with a Tetrahymena ribozyme, altering the sequence of the IGS of the P.carinii ribozyme can result in a marked reduction in tertiary stability of docking the resultant P1 helix into the catalytic core of the ribozyme. Finally, results indicate that RNA targeting strategies which exploit tertiary interactions could have low specificity due to the tolerance of mismatched base pairs.
Figures



Similar articles
-
A Pneumocystis carinii group I intron ribozyme that does not require 2' OH groups on its 5' exon mimic for binding to the catalytic core.Biochemistry. 1997 Dec 9;36(49):15303-14. doi: 10.1021/bi9713097. Biochemistry. 1997. PMID: 9398259
-
Contributions of individual nucleotides to tertiary binding of substrate by a Pneumocystis carinii group I intron.Biochemistry. 2000 Nov 21;39(46):14269-78. doi: 10.1021/bi001345x. Biochemistry. 2000. PMID: 11087376
-
Tertiary interactions with the internal guide sequence mediate docking of the P1 helix into the catalytic core of the Tetrahymena ribozyme.Biochemistry. 1993 Dec 14;32(49):13593-604. doi: 10.1021/bi00212a027. Biochemistry. 1993. PMID: 7504953
-
Mutations in the Tetrahymena ribozyme internal guide sequence: effects on docking of the P1 helix into the catalytic core and correlation with catalytic activity.Biochemistry. 1996 Sep 3;35(35):11493-502. doi: 10.1021/bi960510z. Biochemistry. 1996. PMID: 8784205
-
New approaches to targeting RNA with oligonucleotides: inhibition of group I intron self-splicing.Biopolymers. 2004 Jan;73(1):151-61. doi: 10.1002/bip.10520. Biopolymers. 2004. PMID: 14691946 Review.
Cited by
-
Canonical nucleosides can be utilized by T4 DNA ligase as universal template bases at ligation junctions.Nucleic Acids Res. 2003 Jun 15;31(12):3208-16. doi: 10.1093/nar/gkg415. Nucleic Acids Res. 2003. PMID: 12799448 Free PMC article.
-
Toxic introns and parasitic intein in Coxiella burnetii: legacies of a promiscuous past.J Bacteriol. 2008 Sep;190(17):5934-43. doi: 10.1128/JB.00602-08. Epub 2008 Jul 7. J Bacteriol. 2008. PMID: 18606739 Free PMC article.
-
Splicing and evolution of an unusually small group I intron.Curr Genet. 2008 Oct;54(4):213-22. doi: 10.1007/s00294-008-0213-y. Epub 2008 Sep 6. Curr Genet. 2008. PMID: 18777024
References
-
- Kruger K., Grabowski,P.J., Zaug,A.J., Sands,J., Gottschling,D.E. and Cech,T.R. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell, 31, 147–157. - PubMed
-
- Zaug A.J., Grabowski,P.J. and Cech,T.R. (1983) Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature, 301, 578–583. - PubMed
-
- Zaug A.J., Kent,J.R. and Cech,T.R. (1984) A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science, 224, 574–578. - PubMed
-
- Zaug A.J., Been,M.D. and Cech,T.R. (1986) The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature, 324, 429–433. - PubMed
-
- Zaug A.J. and Cech,T.R. (1986) The intervening sequence RNA of Tetrahymena is an enzyme. Science, 231, 470–475. - PubMed

