Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays
- PMID: 12655013
- PMCID: PMC152799
- DOI: 10.1093/nar/gkg283
Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays
Abstract
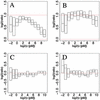
Oligonucleotide microarrays are based on the hybridization of labeled mRNA molecules to short length oligonucleotide probes on a glass surface. Two effects have been shown to affect the raw data: the sequence dependence of the probe hybridization properties and the chemical saturation resulting from surface adsorption processes. We address both issues simultaneously using a physically motivated hybridization model. Based on publicly available calibration data sets, we show that Langmuir adsorption accurately describes GeneChip hybridization, with model parameters that we predict from the sequence composition of the probes. Because these parameters have physical units, we are able to estimate absolute mRNA concentrations in picomolar. Additionally, by accounting for chemical saturation, we substantially reduce the compressive bias of differential expression estimates that normally occurs toward high concentrations.
Figures
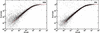
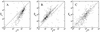


References
-
- Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Accessing genetic information with high-density DNA arrays. Science, 274, 610–614. - PubMed
-
- Naef F., Socci,N. and Magnasco,M. (2002) A study of accuracy and precision in oligonucleotide arrays: extracting more signal at large concentrations. Bioinformatics, 19, 178–184. - PubMed
-
- Naef F., Lim,D.A., Patil,N. and Magnasco,M. (2002) DNA hybridization to mismatched templates: a chip study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys., 65, 040902. - PubMed

