Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth
- PMID: 12657681
- PMCID: PMC6742005
- DOI: 10.1523/JNEUROSCI.23-06-02218.2003
Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth
Abstract
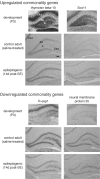
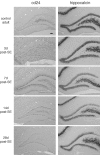
Neurogenesis and axon outgrowth are features shared by normal nervous system development and certain forms of epileptogenesis. This observation has led to the hypothesis that some aspects of normal development and epileptogenesis have common molecular mechanisms. To test this hypothesis, we have used DNA microarray analysis to characterize gene expression in the dentate gyrus and identify genes exhibiting similar patterns of regulation during development and epileptogenesis. Of more than 8000 sequences surveyed, over 600 were regulated during development or epileptogenesis, and 37 of these were either upregulated or downregulated during both processes. In situ hybridization analysis of a subset of these "commonality genes" confirmed the patterns of regulation predicted by the microarray data in most cases and demonstrated various spatial and temporal patterns of commonality gene expression. Of the 25 named commonality genes in which some functional characteristics are known, 11 have been implicated in cell morphology and axon outgrowth or cellular proliferation and fate determination. This enrichment for candidate plasticity-related genes supports the concept that developmental mechanisms contribute to network alterations associated with epileptogenesis and offers a useful strategy for identifying molecules that may play a role in both of these processes.
Figures



References
-
- Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J Comp Neurol. 1990;301:365–381. - PubMed
-
- Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. - PubMed
-
- Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with 3H-thymidine autoradiography. J Comp Neurol. 1980;190:87–114. - PubMed
-
- Braunewell KH, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999;295:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
