The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
- PMID: 12660157
- PMCID: PMC152889
- DOI: 10.1093/emboj/cdg144
The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Abstract
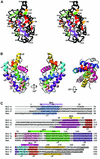
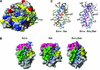
Pro-survival Bcl-2-related proteins, critical regulators of apoptosis, contain a hydrophobic groove targeted for binding by the BH3 domain of the pro-apoptotic BH3-only proteins. The solution structure of the pro-survival protein Bcl-w, presented here, reveals that the binding groove is not freely accessible as predicted by previous structures of pro-survival Bcl-2-like molecules. Unexpectedly, the groove appears to be occluded by the C-terminal residues. Binding and kinetic data suggest that the C-terminal residues of Bcl-w and Bcl-x(L) modulate pro-survival activity by regulating ligand access to the groove. Binding of the BH3-only proteins, critical for cell death initiation, is likely to displace the hydrophobic C-terminal region of Bcl-w and Bcl-x(L). Moreover, Bcl-w does not act only by sequestering the BH3-only proteins. There fore, pro-survival Bcl-2-like molecules probably control the activation of downstream effectors by a mechanism that remains to be elucidated.
Figures



References
-
- Aritomi M., Kunishima,N., Inohara,N., Ishibashi,Y., Ohta,S. and Morikawa,K. (1997) Crystal structure of Rat Bcl-xL. J. Biol. Chem., 272, 27886–27892. - PubMed
-
- Bartels C., Xia,T.H., Billeter,M., Güntert,P. and Wüthrich,K. (1995) The program XEASY for computer-supported nmr spectral-analysis of biological macromolecules. J. Biomol. NMR, 6, 1–10. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D., 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

