Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin
- PMID: 12660172
- PMCID: PMC152887
- DOI: 10.1093/emboj/cdg142
Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin
Abstract
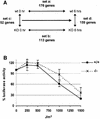
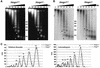
We report that HMGN1, a nucleosome binding protein that destabilizes the higher-order chromatin structure, modulates the repair rate of ultraviolet light (UV)-induced DNA lesions in chromatin. Hmgn1(-/-) mouse embryonic fibroblasts (MEFs) are hypersensitive to UV, and the removal rate of photoproducts from the chromatin of Hmgn1(-/-) MEFs is decreased as compared with the chromatin of Hmgn1(+/+) MEFs; yet, host cell reactivation assays and DNA array analysis indicate that the nucleotide excision repair (NER) pathway in the Hmgn1(-/-) MEFs remains intact. The UV hypersensitivity of Hmgn1(-/-) MEFs could be rescued by transfection with plasmids expressing wild-type HMGN1 protein, but not with plasmids expressing HMGN1 mutants that do not bind to nucleosomes or do not unfold chromatin. Transcriptionally active genes, the main target of the NER pathways in mice, contain HMGN1 protein, and loss of HMGN1 protein reduces the accessibility of transcribed genes to nucleases. By reducing the compaction of the higher-order chromatin structure, HMGN1 facilitates access to UV-damaged DNA sites and enhances the rate of DNA repair in chromatin.
Figures




References
-
- Archer T.K., Lefebvre,P., Wolford,R.G. and Hager,G.L. (1992) Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science, 255, 1573–1576. - PubMed
-
- Balajee A.S. and Bohr,V.A. (2000) Genomic heterogeneity of nucleotide excision repair. Gene, 250, 15–30. - PubMed
-
- Bill C.A., Grochan,B.M., Meyn,R.E., Bohr,V.A. and Tofilon,P.J. (1991) Loss of intragenomic DNA repair heterogeneity with cellular differentiation. J. Biol. Chem., 266, 21821–21826. - PubMed
-
- Bohr V.A. (1991) Gene specific DNA repair. Carcinogenesis, 12, 1983–1992. - PubMed
-
- Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell, 40, 359–369. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

