Interactions between papillomavirus L1 and L2 capsid proteins
- PMID: 12663788
- PMCID: PMC152166
- DOI: 10.1128/jvi.77.8.4818-4826.2003
Interactions between papillomavirus L1 and L2 capsid proteins
Abstract
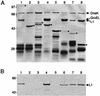
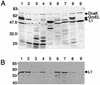
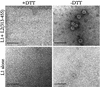
The human papillomavirus (HPV) capsid consists of 360 copies of the major capsid protein, L1, arranged as 72 pentamers on a T=7 icosahedral lattice, with substoichiometric amounts of the minor capsid protein, L2. In order to understand the arrangement of L2 within the HPV virion, we have defined and biochemically characterized a domain of L2 that interacts with L1 pentamers. We utilized an in vivo binding assay involving the coexpression of recombinant HPV type 11 (HPV11) L1 and HPV11 glutathione S-transferase (GST) L2 fusion proteins in Escherichia coli. In this system, L1 forms pentamers, GST=L2 associates with these pentamers, and L1+L2 complexes are subsequently isolated by using the GST tag on L2. The stoichiometry of L1:L2 in purified L1+L2 complexes was 5:1, indicating that a single molecule of L2 interacts with an L1 pentamer. Coexpression of HPV11 L1 with deletion mutants of HPV11 L2 defined an L1-binding domain contained within amino acids 396 to 439 near the carboxy terminus of L2. L2 proteins from eight different human and animal papillomavirus serotypes were tested for their ability to interact with HPV11 L1. This analysis targeted a hydrophobic region within the L1-binding domain of L2 as critical for L1 binding. Introduction of negative charges into this hydrophobic region by site-directed mutagenesis disrupted L1 binding. L1-L2 interactions were not significantly disrupted by treatment with high salt concentrations (2 M NaCl), weak detergents, and urea concentrations of up to 2 M, further indicating that L1 binding by this domain is mediated by strong hydrophobic interactions. L1+L2 protein complexes were able to form virus-like particles in vitro at pH 5.2 and also at pH 6.8, a pH that is nonpermissive for assembly of L1 protein alone. Thus, L1/L2 interactions are primarily hydrophobic, encompass a relatively short stretch of amino acids, and have significant effects upon in vitro assembly.
Figures






References
-
- Anonymous. 1997. Phylogenetic tree, p. 1-3. In G. Myers, C. Baker, K. Münger, F. Sverdrup, A. McBride, H.-U. Bernard, and J. Meissner (ed.), Human papillomaviruses. Los Alamos National Laboratory Theoretical Biology and Biophysics Group, Los Alamos, N.Mex.
-
- Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. - PubMed
-
- Cain, J. M., and M. K. Howett. 2000. Preventing cervical cancer. Science 288:1753-1755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

