Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function
- PMID: 12665577
- PMCID: PMC152559
- DOI: 10.1128/MCB.23.8.2762-2777.2003
Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function
Abstract
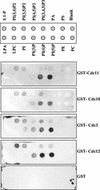
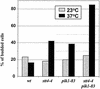
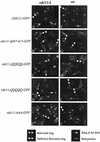
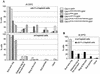
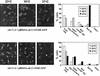
The septins are a family of cytoskeletal proteins present in animal and fungal cells. They were first identified for their essential role in cytokinesis, but more recently, they have been found to play an important role in many cellular processes, including bud site selection, chitin deposition, cell compartmentalization, and exocytosis. Septin proteins self-associate into filamentous structures that, in yeast cells, form a cortical ring at the mother bud neck. Members of the septin family share common structural domains: a GTPase domain in the central region of the protein, a stretch of basic residues at the amino terminus, and a predicted coiled-coil domain at the carboxy terminus. We have studied the role of each domain in the Saccharomyces cerevisiae septin Cdc11 and found that the three domains are responsible for distinct and sometimes overlapping functions. All three domains are important for proper localization and function in cytokinesis and morphogenesis. The basic region was found to bind the phosphoinositides phosphatidylinositol 4-phosphate and phosphatidylinositol 5-phosphate. The coiled-coil domain is important for interaction with Cdc3 and Bem4. The GTPase domain is involved in Cdc11-septin interaction and targeting to the mother bud neck. Surprisingly, GTP binding appears to be dispensable for Cdc11 function, localization, and lipid binding. Thus, we find that septins are multifunctional proteins with specific domains involved in distinct molecular interactions required for assembly, localization, and function within the cell.
Figures







References
-
- Audhya, A., and S. D. Emr. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2:593-605. - PubMed
-
- Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
