A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo
- PMID: 12665580
- PMCID: PMC152555
- DOI: 10.1128/MCB.23.8.2800-2820.2003
A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo
Abstract
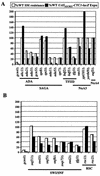
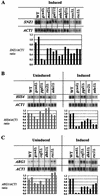
Transcriptional activators interact with multisubunit coactivators that modify chromatin structure or recruit the general transcriptional machinery to their target genes. Budding yeast cells respond to amino acid starvation by inducing an activator of amino acid biosynthetic genes, Gcn4p. We conducted a comprehensive analysis of viable mutants affecting known coactivator subunits from the Saccharomyces Genome Deletion Project for defects in activation by Gcn4p in vivo. The results confirm previous findings that Gcn4p requires SAGA, SWI/SNF, and SRB mediator (SRB/MED) and identify key nonessential subunits of these complexes required for activation. Among the numerous histone acetyltransferases examined, only that present in SAGA, Gcn5p, was required by Gcn4p. We also uncovered a dependence on CCR4-NOT, RSC, and the Paf1 complex. In vitro binding experiments suggest that the Gcn4p activation domain interacts specifically with CCR4-NOT and RSC in addition to SAGA, SWI/SNF, and SRB/MED. Chromatin immunoprecipitation experiments show that Mbf1p, SAGA, SWI/SNF, SRB/MED, RSC, CCR4-NOT, and the Paf1 complex all are recruited by Gcn4p to one of its target genes (ARG1) in vivo. We observed considerable differences in coactivator requirements among several Gcn4p-dependent promoters; thus, only a subset of the array of coactivators that can be recruited by Gcn4p is required at a given target gene in vivo.
Figures







References
-
- Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
