The carboxy terminus of the small subunit of TFIIE regulates the transition from transcription initiation to elongation by RNA polymerase II
- PMID: 12665589
- PMCID: PMC152561
- DOI: 10.1128/MCB.23.8.2914-2926.2003
The carboxy terminus of the small subunit of TFIIE regulates the transition from transcription initiation to elongation by RNA polymerase II
Abstract
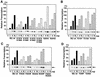
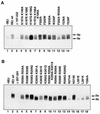
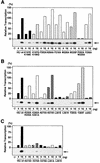
The general transcription factor TFIIE plays essential roles in both transcription initiation and the transition from initiation to elongation. Previously, we systematically deleted the structural motifs and characteristic sequences of the small subunit of human TFIIE (hTFIIE beta) to map its functional regions. Here we introduced point mutations into two regions located near the carboxy terminus of hTFIIE beta and identified the functionally essential amino acid residues that bind to RNA polymerase II (Pol II), the general transcription factors, and single-stranded DNA. Although most residues identified were essential for transcription initiation, use of an in vitro transcription assay with a linearized template revealed that several residues in the carboxy-terminal helix-loop region are crucially involved in the transition stage. Mutations in these residues also affected the ability of hTFIIE beta to stimulate TFIIH-mediated phosphorylation of the carboxy-terminal heptapeptide repeats of the largest subunit of Pol II. Furthermore, these mutations conspicuously augmented the binding of hTFIIE beta to the p44 subunit of TFIIH. The antibody study indicated that they thus altered the conformation of one side of TFIIH, consisting of p44, XPD, and Cdk-activating kinase subunits, that is essential for the transition stage. This is an important clue for elucidating the molecular mechanisms involved in the transition stage.
Figures







References
-
- Chang, W.-H., and R. D. Kornberg. 2000. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell 102:609-613. - PubMed
-
- Coin, F., J.-C. Marinoni, C. Rodolfo, S. Fribourg, A. M. Pedrini, and J.-M. Egly. 1998. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20:184-188. - PubMed
-
- Douziech, M., F. Coin, J. M. Chipoulet, Y. Arai, Y. Ohkuma, J. M. Egly, and B. Coulombe. 2000. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell. Biol. 20:8168-8177. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
