Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing
- PMID: 12665616
- PMCID: PMC153610
- DOI: 10.1073/pnas.0630618100
Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing
Abstract
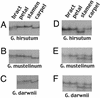
Most eukaryotes have genomes that exhibit high levels of gene redundancy, much of which seems to have arisen from one or more cycles of genome doubling. Polyploidy has been particularly prominent during flowering plant evolution, yielding duplicated genes (homoeologs) whose expression may be retained or lost either as an immediate consequence of polyploidization or on an evolutionary timescale. Expression of 40 homoeologous gene pairs was assayed by cDNA-single-stranded conformation polymorphism in natural (1- to 2-million-yr-old) and synthetic tetraploid cotton (Gossypium) to determine whether homoeologous gene pairs are expressed at equal levels after polyploid formation. Silencing or unequal expression of one homoeolog was documented for 10 of 40 genes examined in ovules of Gossypium hirsutum. Assays of homoeolog expression in 10 organs revealed variable expression levels and silencing, depending on the gene and organ examined. Remarkably, silencing and biased expression of some gene pairs are reciprocal and developmentally regulated, with one homoeolog showing silencing in some organs and the other being silenced in other organs, suggesting rapid subfunctionalization. Duplicate gene expression was examined in additional natural polyploids to characterize the pace at which expression alteration evolves. Analysis of a synthetic tetraploid revealed homoeolog expression and silencing patterns that sometimes mirrored those of the natural tetraploid. Both long-term and immediate responses to polyploidization were implicated. Data suggest that some silencing events are epigenetically induced during the allopolyploidization process.
Figures


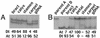
Comment in
-
What happens to genes in duplicated genomes.Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4369-71. doi: 10.1073/pnas.0831050100. Epub 2003 Apr 7. Proc Natl Acad Sci U S A. 2003. PMID: 12682287 Free PMC article. No abstract available.
References
-
- Ohno S. Evolution by Gene Duplication. New York: Springer; 1970.
-
- Gu X, Wang Y, Gu J. Nat Genet. 2002;31:205–209. - PubMed
-
- McLysaght A, Hokamp K, Wolfe K H. Nat Genet. 2002;31:200–204. - PubMed
-
- Wolfe K H, Shields D C. Nature. 1997;387:708–713. - PubMed
-
- Wendel J F. Plant Mol Biol. 2000;42:225–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

