Dynamics of DNA molecules in a membrane channel probed by active control techniques
- PMID: 12668445
- PMCID: PMC1302803
- DOI: 10.1016/S0006-3495(03)75042-5
Dynamics of DNA molecules in a membrane channel probed by active control techniques
Abstract
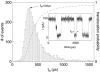
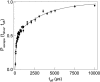
The dynamics of single-stranded DNA in an alpha-Hemolysin protein pore was studied at the single-molecule level. The escape time for DNA molecules initially drawn into the pore was measured in the absence of an externally applied electric field. These measurements revealed two well-separated timescales, one of which is surprisingly long (on the order of milliseconds). We characterized the long timescale as being associated with the binding and unbinding of DNA from the pore. We have also found that a transmembrane potential as small as 20 mV strongly biased the escape of DNA from the pore. These experiments have been made possible due to the development of a feedback control system, allowing the rapid modulation of the applied force on individual DNA molecules while inside the pore.
Figures





References
-
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell. Garland Publishing, New York.
-
- Bezrukov, S. M. 2000. Ion channels as molecular Coulter counters to probe metabolite transport. J. Mem. Biol. 174:1–13. - PubMed
-
- Chuang, J., Y. Kantor, and M. Kardar. 2002. Anomalous dynamics of translocation. Phys. Rev. E. 65:011802. - PubMed
-
- Colquhoun, D., and A. G. Hawkes. 1995. The principles of the stochastic interpretation of ion-channel mechanics. In Single-Channel Recording. B. Sakmann and E. Neher, editors. Plenum Press, New York.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

