Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis
- PMID: 12670965
- PMCID: PMC152626
- DOI: 10.1128/JB.185.8.2418-2431.2003
Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis
Abstract
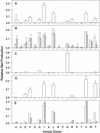
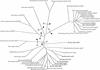
The conversion of ketomethiobutyrate to methionine has been previously examined in a number of organisms, wherein the aminotransferases responsible for the reaction have been found to be members of the Ia subfamily (L. C. Berger, J. Wilson, P. Wood, and B. J. Berger, J. Bacteriol. 183:4421-4434, 2001). The genome of Bacillus subtilis has been found to contain no subfamily Ia aminotransferase sequences. Instead, the analogous enzymes in B. subtilis were found to be members of the If subfamily. These putative aspartate aminotransferases, the yugH, ywfG, ykrV, aspB, and patA gene products, have been cloned, expressed, and characterized for methionine regeneration activity. Only YkrV was able to convert ketomethiobutyrate to methionine, and it catalyzed the reaction only when glutamine was used as amino donor. In contrast, subcellular homogenates of B. subtilis and Bacillus cereus utilized leucine, isoleucine, valine, alanine, phenylalanine, and tyrosine as effective amino donors. The two putative branched-chain aminotransferase genes in B. subtilis, ybgE and ywaA, were also cloned, expressed, and characterized. Both gene products effectively transaminated branched-chain amino acids and ketoglutarate, but only YbgE converted ketomethiobutyrate to methionine. The amino donor preference for methionine regeneration by YbgE was found to be leucine, isoleucine, valine, phenylalanine, and tyrosine. The B. subtilis ybgE gene is a member of the family III of aminotransferases and falls in a subfamily designated here IIIa. Examination of B. cereus and Bacillus anthracis genome data found that there were no subfamily IIIa homologues in these organisms. In both B. cereus and B. anthracis, two putative branched-chain aminotransferases and two putative D-amino acid aminotransferases were discovered as members of subfamily IIIb. These four sequences were cloned from B. cereus, expressed, and characterized. Only the gene product from the sequence designated Bc-BCAT2 was found to convert ketomethiobutyrate to methionine, with an amino donor preference of leucine, isoleucine, valine, phenylalanine, and tyrosine. The B. anthracis homologue of Bc-BCAT2 was also cloned, expressed, and characterized and was found to be identical in activity. The aminooxy compound canaline was found to be an uncompetitive inhibitor of B. subtilis YbgE and also inhibited growth of B. subtilis and B. cereus in culture.
Figures







References
-
- Backlund, P. S., Jr., and R. A. Smith. 1981. Methionine synthesis from 5′-methylthioadenosine in rat liver. J. Biol. Chem. 256:1533-1535. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

