Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942
- PMID: 12670983
- PMCID: PMC152603
- DOI: 10.1128/JB.185.8.2582-2591.2003
Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942
Abstract
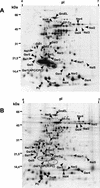
The transcription factor of the cyclic AMP receptor protein/FNR family, NtcA, and the P(II) signaling protein play central roles in global nitrogen control in cyanobacteria. A dependence on P(II) for NtcA-regulated transcription, however, has not been observed. In the present investigation, we examined alterations in gene expression following nitrogen deprivation in Synechococcus elongatus strain PCC 7942 and specifically the roles of NtcA and P(II). Global changes in de novo protein synthesis following combined-nitrogen deprivation were visualized by in vivo [(35)S]methionine labeling and two-dimensional polyacrylamide gel electrophoresis analysis. Nearly all proteins whose synthesis responded specifically to combined-nitrogen deprivation in wild-type cells of S. elongatus failed to respond in P(II)- and NtcA-deficient mutants. One of the proteins whose synthesis was down-regulated in a P(II)- and NtcA-dependent manner was RbcS, the small subunit of RubisCO. Quantification of its mRNA revealed that the abundance of the rbcLS transcript following combined-nitrogen deprivation rapidly declined in wild-type cells but not in P(II) and NtcA mutant cells. To investigate further the relationship between P(II) and NtcA, fusions of the promotorless luxAB reporter genes to the NtcA-regulated glnB gene were constructed and these constructs were used to transform wild-type cells and P(II)(-) and NtcA(-) mutants. Determination of bioluminescence under different growth conditions showed that NtcA represses gene expression in the presence of ammonium in a P(II)-independent manner. By contrast, NtcA-dependent activation of glnB expression following combined-nitrogen deprivation was impaired in the absence of P(II). Together, these results suggest that under conditions of combined-nitrogen deprivation, the regulation of NtcA-dependent gene expression requires the P(II) signal transduction protein.
Figures






References
-
- Allen, M. M., and A. J. Smith. 1969. Nitrogen chlorosis in blue-green algae. Arch. Microbiol. 69:114-120. - PubMed
-
- Andersson, C. R., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. Min, and S. S. Golden. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305:527-542. - PubMed
-
- Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

