Frequency domain analysis of noise in autoregulated gene circuits
- PMID: 12671069
- PMCID: PMC404696
- DOI: 10.1073/pnas.0736140100
Frequency domain analysis of noise in autoregulated gene circuits
Abstract
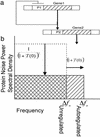
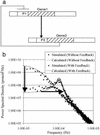
We describe a frequency domain technique for the analysis of intrinsic noise within negatively autoregulated gene circuits. This approach is based on the transfer function around the feedback loop (loop transmission) and the equivalent noise bandwidth of the system. The loop transmission, T, is shown to be a determining factor of the dynamics and the noise behavior of autoregulated gene circuits, and this T-based technique provides a simple and flexible method for the analysis of noise arising from any source within the gene circuit. We show that negative feedback not only reduces the variance of the noise in the protein concentration, but also shifts this noise to higher frequencies where it may have a negligible effect on the noise behavior of following gene circuits within a cascade. This predicted effect is demonstrated through the exact stochastic simulation of a two-gene cascade. The analysis elucidates important aspects of gene circuit structure that control functionality, and may provide some insights into selective pressures leading to this structure. The resulting analytical relationships have a simple form, making them especially useful as synthetic gene circuit design equations. With the exception of the linearization of Hill kinetics, this technique is general and may be applied to the analysis or design of networks of higher complexity. This utility is demonstrated through the exact stochastic simulation of an autoregulated two-gene cascade operating near instability.
Figures



References
-
- Rao C. V., Wolf, D. M. & Arkin, A. P. (2002) Nature 420, 231-237. - PubMed
-
- De Jong H. (2002) J. Comput. Biol. 9, 67-103. - PubMed
-
- Endy D. & Brent, R. (2001) Nature 409, 391-395. - PubMed
-
- Hasty J., McMillen, D., Isaacs, F. & Collins, J. J. (2001) Nat. Rev. Genet. 2, 268-279. - PubMed
-
- McAdams H. H. & Arkin, A. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 199-224. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

