Site-specific contributions to the pH dependence of protein stability
- PMID: 12671071
- PMCID: PMC404695
- DOI: 10.1073/pnas.0736600100
Site-specific contributions to the pH dependence of protein stability
Abstract
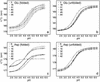
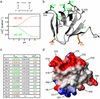
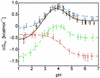
Understanding protein stability is a significant challenge requiring characterization of interactions within both folded and unfolded states. Of these, electrostatic interactions influence ionization equilibria of acidic and basic groups and diversify their pK(a) values. The pH dependence of the thermodynamic stability (Delta G(FU)) of a protein arises as a consequence of differential pK(a) values between folded and unfolded states. Previous attempts to calculate pH-dependent contributions to stability have been limited by the lack of experimental unfolded state pK(a) values. Using recently developed NMR spectroscopic methods, we have determined residue-specific pK(a) values for a thermodynamically unstable Src homology 3 domain in both states, enabling the calculation of the pH dependence of stability based on simple analytical expressions. The calculated pH stability profile obtained agrees very well with experiment, unlike profiles derived from two current models of electrostatic interactions within unfolded states. Most importantly, per-residue contributions to the pH dependence of Delta G(FU) derived from the data provide insights into specific electrostatic interactions in both the folded and unfolded states and their roles in protein stability. These interactions include a hydrogen bond between the Asp-8 side-chain and the Lys-21 backbone amide group in the folded state, which represents a highly conserved interaction in Src homology 3 domains.
Figures

References
-
- Wong K. B., Freund, S. M. & Fersht, A. R. (1996) J. Mol. Biol. 259, 805-818. - PubMed
-
- Mok Y. K., Kay, C. M., Kay, L. E. & Forman-Kay, J. D. (1999) J. Mol. Biol. 289, 619-638. - PubMed
-
- Shortle D. & Ackerman, M. S. (2001) Science 293, 487-489. - PubMed
-
- Klein-Seetharaman J., Oikawa, M., Grimshaw, S. B., Wirmer, J., Duchardt, E., Ueda, T., Imoto, T., Smith, L. J., Dobson, C. M. & Schwalbe, H. (2002) Science 295, 1719-1722. - PubMed
-
- Crowhurst K. A., Tollinger, M. & Forman-Kay, J. D. (2002) J. Mol. Biol. 322, 163-178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

