Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis
- PMID: 12671079
- PMCID: PMC152331
- DOI: 10.1105/tpc.009308
Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis
Erratum in
- Plant Cell. 2003 Jul;15(7):1683
Abstract
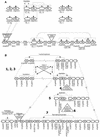
The Arabidopsis genome contains approximately 200 genes that encode proteins with similarity to the nucleotide binding site and other domains characteristic of plant resistance proteins. Through a reiterative process of sequence analysis and reannotation, we identified 149 NBS-LRR-encoding genes in the Arabidopsis (ecotype Columbia) genomic sequence. Fifty-six of these genes were corrected from earlier annotations. At least 12 are predicted to be pseudogenes. As described previously, two distinct groups of sequences were identified: those that encoded an N-terminal domain with Toll/Interleukin-1 Receptor homology (TIR-NBS-LRR, or TNL), and those that encoded an N-terminal coiled-coil motif (CC-NBS-LRR, or CNL). The encoded proteins are distinct from the 58 predicted adapter proteins in the previously described TIR-X, TIR-NBS, and CC-NBS groups. Classification based on protein domains, intron positions, sequence conservation, and genome distribution defined four subgroups of CNL proteins, eight subgroups of TNL proteins, and a pair of divergent NL proteins that lack a defined N-terminal motif. CNL proteins generally were encoded in single exons, although two subclasses were identified that contained introns in unique positions. TNL proteins were encoded in modular exons, with conserved intron positions separating distinct protein domains. Conserved motifs were identified in the LRRs of both CNL and TNL proteins. In contrast to CNL proteins, TNL proteins contained large and variable C-terminal domains. The extant distribution and diversity of the NBS-LRR sequences has been generated by extensive duplication and ectopic rearrangements that involved segmental duplications as well as microscale events. The observed diversity of these NBS-LRR proteins indicates the variety of recognition molecules available in an individual genotype to detect diverse biotic challenges.
Figures







Comment in
-
Resistance rodeo: rounding up the full complement of Arabidopsis NBS-LRR genes.Plant Cell. 2003 Apr;15(4):806-7. doi: 10.1105/tpc.150470. Plant Cell. 2003. PMID: 12671078 Free PMC article. No abstract available.
References
-
- Aarts, M.G., te Lintel Hekkert, B., Holub, E.B., Beynon, J.L., Stiekema, W.J., and Pereira, A. (1998). Identification of R gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 11, 251–258. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aravind, L., Dixit, V.M., and Koonin, E.V. (1999). The domains of death: Evolution of the apoptosis machinery. Trends Biochem. Sci. 24, 47–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

