Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling
- PMID: 12671089
- PMCID: PMC152340
- DOI: 10.1105/tpc.010538
Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling
Abstract
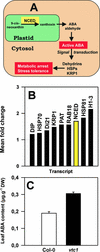
Vitamin C deficiency in the Arabidopsis mutant vtc1 causes slow growth and late flowering. This is not attributable to changes in photosynthesis or increased oxidative stress. We have used the vtc1 mutant to provide a molecular signature for vitamin C deficiency in plants. Using statistical analysis, we show that 171 genes are expressed differentially in vtc1 compared with the wild type. Many defense genes are activated, particularly those that encode pathogenesis-related proteins. Furthermore, transcript changes indicate that growth and development are constrained in vtc1 by the modulation of abscisic acid signaling. Abscisic acid contents are significantly higher in vtc1 than in the wild type. Key features of the molecular signature of ascorbate deficiency can be reversed by incubating vtc1 leaf discs in ascorbate. This finding provides evidence that many of the observed effects on transcript abundance in vtc1 result from ascorbate deficiency. Hence, through modifying gene expression, vitamin C contents not only act to regulate defense and survival but also act via phytohormones to modulate plant growth under optimal conditions.
Figures







References
-
- Agrawal, G.K., Rakwal, R., Jwa, N.S., and Agrawal, V.P. (2001). Signaling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: A model illustrating components participating during defence/stress response. Plant Physiol. Biochem. 39, 1095–1103.
-
- Arrigoni, O., and de Tullio, M.C. (2000). The role of ascorbic acid in cell metabolism: Between gene-directed functions and unpredictable chemical reactions. J. Plant Physiol. 157, 481–488.
-
- Ascenzi, R., and Gantt, S. (1997). A drought-stress inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34, 629–641. - PubMed
-
- Bianco, J., and Dalstein, L. (1999). Abscisic acid in needles of Pinus cembra in relation to ozone exposure. Tree Physiol. 19, 787–791. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

