Processing, localization, and requirement of human separase for normal anaphase progression
- PMID: 12672959
- PMCID: PMC153597
- DOI: 10.1073/pnas.0730733100
Processing, localization, and requirement of human separase for normal anaphase progression
Abstract
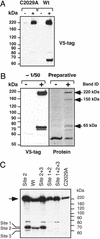
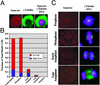
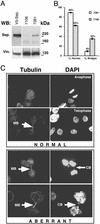
In all eukaryotes, anaphase is triggered by the activation of a protease called separase. Once activated, separase cleaves a subunit of cohesin, a complex that links replicated chromatids before anaphase. Separase and cohesin are conserved from yeasts to humans. Although the machinery for dissolving sister cohesion is conserved, the regulation of this process appears to be more complex in higher eukaryotes than in yeast. Here we report the cloning of full-length human separase cDNA and the characterization of the encoded protein. Human separase was observed at the poles of the mitotic spindle until anaphase, at which time its association with the mitotic spindle was abruptly lost. The dynamic pattern of localization of human separase during cell cycle progression differs from that of fungal separases. Human separase also appears to undergo an autocatalytic processing on anaphase entry. The processed forms of human separase were isolated and the identity of the cleavage sites was determined by N-terminal sequencing and site-directed mutagenesis. The processed catalytic domain was found to be stably associated with the processed N-terminal fragment. Finally, by depletion of endogenous separase with antisense oligonucleotides, we report direct evidence that separase is required for high-fidelity chromosome separation in human cells.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

