Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring
- PMID: 12672969
- PMCID: PMC153575
- DOI: 10.1073/pnas.0330734100
Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring
Abstract
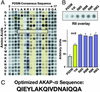
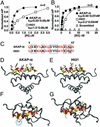
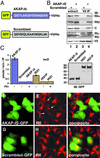
Compartmentalization of the cAMP-dependent protein kinase (PKA) is coordinated through association with A-kinase anchoring proteins (AKAPs). A defining characteristic of most AKAPs is a 14- to 18-aa sequence that binds to the regulatory subunits (RI or RII) of the kinase. Cellular delivery of peptides to these regions disrupts PKA anchoring and has been used to delineate a physiological role for AKAPs in the facilitation of certain cAMP-responsive events. Here, we describe a bioinformatic approach that yields an RII-selective peptide, called AKAP-in silico (AKAP-IS), that binds RII with a K(d) of 0.4 nM and binds RI with a K(d) of 277 nM. AKAP-IS associates with the type II PKA holoenzyme inside cells and displaces the kinase from natural anchoring sites. Electrophysiological recordings indicate that perfusion of AKAP-IS evokes a more rapid and complete attenuation of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor currents than previously described anchoring inhibitor peptides. Thus, computer-based and peptide array screening approaches have generated a reagent that binds PKA with higher affinity than previously described AKAPs.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

