Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract
- PMID: 12676681
- PMCID: PMC154805
- DOI: 10.1128/AEM.69.4.2044-2051.2003
Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract
Abstract
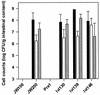
Lactobacilli are common inhabitants of the gastrointestinal tracts of mammals and have received considerable attention due to their putative health-promoting properties. Little is known about the traits that enhance the ability of these bacteria to inhabit the gastrointestinal tract. In this paper we describe the development and application of a strategy based on in vivo expression technology (IVET) that enables detection of Lactobacillus reuteri genes specifically induced in the murine gut. A plasmid-based system was constructed containing 'ermGT (which confers lincomycin resistance) as the primary reporter gene for selection of promoters active in the gastrointestinal tract of mice treated with lincomycin. A second reporter gene, 'bglM (beta-glucanase), allowed differentiation between constitutive and in vivo inducible promoters. The system was successfully tested in vitro and in vivo by using a constitutive promoter. Application of the IVET system with chromosomal DNA of L. reuteri 100-23 and reconstituted lactobacillus-free mice revealed three genes induced specifically during colonization. Two of the sequences showed homology to genes encoding xylose isomerase (xylA) and peptide methionine sulfoxide reductase (msrB), which are involved in nutrient acquisition and stress responses, respectively. The third locus showed homology to the gene encoding a protein whose function is not known. Our IVET system has the potential to identify genes of lactobacilli that have not previously been functionally characterized but which may be essential for growth of these bacteria in the gastrointestinal ecosystem.
Figures
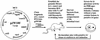

References
-
- Benes, V., Z. Hostomsky, L. Arnold, and V. Paces. 1993. M13 and pUC vectors with new unique restriction sites for cloning. Gene 130:151-152. - PubMed
-
- Borriss, R., K. Buettner, and P. Maentsaelae. 1990. Structure of the beta-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other beta-glucanases. Mol. Gen. Genet. 222:278-283. - PubMed
-
- Brot, N., and H. Weissbach. 2000. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers 55:288-296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

